Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
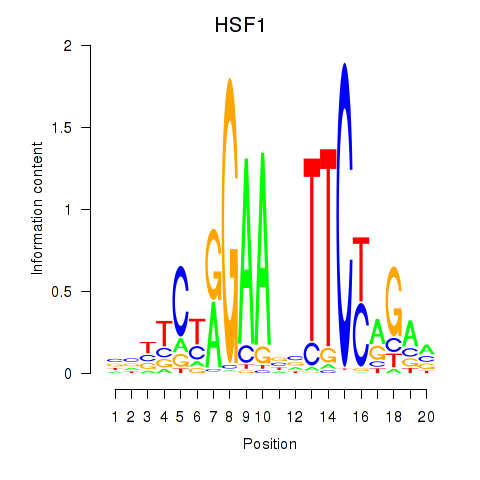
Results for HSF1
Z-value: 0.58
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF1
|
ENSG00000185122.6 | heat shock transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg19_v2_chr8_+_145515263_145515299 | -0.13 | 8.1e-01 | Click! |
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_113735575 | 0.66 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr19_-_6720686 | 0.53 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr2_+_228678550 | 0.50 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr1_-_153363452 | 0.49 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr20_+_43803517 | 0.44 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr11_+_69061594 | 0.43 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr19_+_3762703 | 0.41 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr10_+_15001430 | 0.40 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr11_-_105010320 | 0.39 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr19_+_782755 | 0.36 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr2_+_132044330 | 0.35 |
ENST00000416266.1
|
CYP4F31P
|
cytochrome P450, family 4, subfamily F, polypeptide 31, pseudogene |
| chr21_+_42733870 | 0.33 |
ENST00000330714.3
ENST00000436410.1 ENST00000435611.1 |
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr17_-_54911250 | 0.29 |
ENST00000575658.1
ENST00000397861.2 |
C17orf67
|
chromosome 17 open reading frame 67 |
| chr5_-_150466692 | 0.27 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr4_+_76481258 | 0.25 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr17_+_45331184 | 0.25 |
ENST00000559488.1
ENST00000571680.1 ENST00000435993.2 |
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| chr6_-_38607628 | 0.24 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr7_-_139763521 | 0.24 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr16_-_2770216 | 0.23 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr3_-_10334617 | 0.22 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.8 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr2_+_40973618 | 0.22 |
ENST00000420187.1
|
AC007317.1
|
AC007317.1 |
| chr7_-_150020814 | 0.21 |
ENST00000477871.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr19_+_3762645 | 0.21 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr2_-_191878162 | 0.20 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr5_+_131630117 | 0.19 |
ENST00000200652.3
|
SLC22A4
|
solute carrier family 22 (organic cation/zwitterion transporter), member 4 |
| chr5_-_150467221 | 0.19 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr4_+_57396766 | 0.19 |
ENST00000512175.2
|
THEGL
|
theg spermatid protein-like |
| chr19_-_2085323 | 0.19 |
ENST00000591638.1
|
MOB3A
|
MOB kinase activator 3A |
| chr1_-_183538319 | 0.18 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr19_+_49055332 | 0.18 |
ENST00000201586.2
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr6_-_29648887 | 0.18 |
ENST00000376883.1
|
ZFP57
|
ZFP57 zinc finger protein |
| chr2_+_102624977 | 0.18 |
ENST00000441002.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chr2_-_220142892 | 0.17 |
ENST00000427737.1
|
TUBA4A
|
tubulin, alpha 4a |
| chr3_-_10334585 | 0.17 |
ENST00000430179.1
ENST00000449238.2 ENST00000437422.2 ENST00000287656.7 ENST00000457360.1 ENST00000439975.2 ENST00000446937.2 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr6_-_32812420 | 0.17 |
ENST00000374881.2
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr6_-_31782813 | 0.16 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr15_-_86338134 | 0.16 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr11_-_102714534 | 0.16 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr1_+_92683467 | 0.16 |
ENST00000370375.3
ENST00000370373.2 |
C1orf146
|
chromosome 1 open reading frame 146 |
| chr7_+_150020363 | 0.16 |
ENST00000359623.4
ENST00000493307.1 |
LRRC61
|
leucine rich repeat containing 61 |
| chr11_-_47870091 | 0.16 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr20_-_43753104 | 0.16 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr2_-_191878681 | 0.16 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr19_+_56905024 | 0.15 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr2_-_191878874 | 0.15 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chrX_-_100662881 | 0.15 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr14_+_101293687 | 0.15 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr14_-_81425828 | 0.15 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr1_+_212782012 | 0.15 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr2_-_238499131 | 0.14 |
ENST00000538644.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr6_-_112081113 | 0.14 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr14_-_75083313 | 0.14 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr15_+_67390920 | 0.14 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr20_+_2633269 | 0.14 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein |
| chr3_+_112930946 | 0.14 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr12_+_51632666 | 0.14 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr8_+_119294456 | 0.14 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr5_-_56778635 | 0.13 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr1_+_202830876 | 0.13 |
ENST00000456105.2
|
RP11-480I12.7
|
RP11-480I12.7 |
| chr21_+_35445811 | 0.13 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr1_+_212738676 | 0.13 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr3_-_120400960 | 0.13 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr2_-_191885686 | 0.13 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr19_-_55791563 | 0.13 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr8_+_142402089 | 0.13 |
ENST00000521578.1
ENST00000520105.1 ENST00000523147.1 |
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr19_+_8740061 | 0.13 |
ENST00000593792.1
|
CTD-2586B10.1
|
CTD-2586B10.1 |
| chr18_-_71815051 | 0.13 |
ENST00000582526.1
ENST00000419743.2 |
FBXO15
|
F-box protein 15 |
| chrX_-_128782722 | 0.13 |
ENST00000427399.1
|
APLN
|
apelin |
| chr1_-_27682962 | 0.13 |
ENST00000486046.1
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr18_-_71814999 | 0.12 |
ENST00000269500.5
|
FBXO15
|
F-box protein 15 |
| chr11_+_809961 | 0.12 |
ENST00000530797.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr11_+_94706804 | 0.12 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr16_+_2016821 | 0.12 |
ENST00000569210.2
ENST00000569714.1 |
RNF151
|
ring finger protein 151 |
| chr6_-_161695074 | 0.12 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr2_+_58655461 | 0.12 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr2_+_223162866 | 0.12 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chr16_-_57831914 | 0.12 |
ENST00000421376.2
|
KIFC3
|
kinesin family member C3 |
| chr16_-_12184159 | 0.12 |
ENST00000312019.2
|
RP11-276H1.3
|
RP11-276H1.3 |
| chr11_+_34645791 | 0.12 |
ENST00000529527.1
ENST00000531728.1 ENST00000525253.1 |
EHF
|
ets homologous factor |
| chr19_+_35417939 | 0.12 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr7_+_141478242 | 0.12 |
ENST00000247881.2
|
TAS2R4
|
taste receptor, type 2, member 4 |
| chr6_-_32811771 | 0.11 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr2_-_43266680 | 0.11 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr22_-_50708781 | 0.11 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr22_-_36556821 | 0.11 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chrX_+_77359726 | 0.11 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr12_+_120933859 | 0.11 |
ENST00000242577.6
ENST00000548214.1 ENST00000392508.2 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr19_+_49458107 | 0.11 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr12_+_48147699 | 0.11 |
ENST00000548498.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr11_+_60145948 | 0.11 |
ENST00000300184.3
ENST00000358246.1 |
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr4_-_170192000 | 0.11 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr5_+_40909354 | 0.11 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr2_+_232826211 | 0.11 |
ENST00000325385.7
ENST00000360410.4 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr17_+_77019030 | 0.11 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_-_150754935 | 0.10 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr19_+_8429031 | 0.10 |
ENST00000301455.2
ENST00000541807.1 ENST00000393962.2 |
ANGPTL4
|
angiopoietin-like 4 |
| chr9_+_74764278 | 0.10 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr6_+_10694900 | 0.10 |
ENST00000379568.3
|
PAK1IP1
|
PAK1 interacting protein 1 |
| chr5_-_141016382 | 0.10 |
ENST00000523088.1
ENST00000305264.3 |
HDAC3
|
histone deacetylase 3 |
| chr7_+_157129660 | 0.10 |
ENST00000429029.2
ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr19_-_55672037 | 0.10 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr19_+_45174994 | 0.10 |
ENST00000403660.3
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr7_+_2393714 | 0.10 |
ENST00000431643.1
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr6_+_161123270 | 0.10 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr6_-_161695042 | 0.10 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr19_+_35417844 | 0.10 |
ENST00000601957.1
|
ZNF30
|
zinc finger protein 30 |
| chr17_+_73996987 | 0.10 |
ENST00000588812.1
ENST00000448471.1 |
CDK3
|
cyclin-dependent kinase 3 |
| chr17_+_75452447 | 0.10 |
ENST00000591472.2
|
SEPT9
|
septin 9 |
| chr17_-_39928106 | 0.10 |
ENST00000540235.1
|
JUP
|
junction plakoglobin |
| chr6_+_44215603 | 0.10 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr4_-_154681108 | 0.10 |
ENST00000508248.1
ENST00000274068.4 |
RNF175
|
ring finger protein 175 |
| chr3_+_52245458 | 0.10 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr6_+_30294612 | 0.10 |
ENST00000440271.1
ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39
|
tripartite motif containing 39 |
| chr8_-_49533521 | 0.10 |
ENST00000523038.1
|
RP11-567J20.1
|
RP11-567J20.1 |
| chr19_-_2739992 | 0.10 |
ENST00000545664.1
ENST00000589363.1 ENST00000455372.2 |
SLC39A3
|
solute carrier family 39 (zinc transporter), member 3 |
| chr1_-_155948890 | 0.10 |
ENST00000471589.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr6_-_41909191 | 0.10 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr2_-_241831424 | 0.10 |
ENST00000402775.2
ENST00000307486.8 |
C2orf54
|
chromosome 2 open reading frame 54 |
| chr14_-_21493123 | 0.10 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr1_+_67773527 | 0.10 |
ENST00000541374.1
ENST00000544434.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr5_-_171615315 | 0.09 |
ENST00000176763.5
|
STK10
|
serine/threonine kinase 10 |
| chr9_-_97090926 | 0.09 |
ENST00000335456.7
ENST00000253262.4 ENST00000341207.4 |
NUTM2F
|
NUT family member 2F |
| chr7_-_100860851 | 0.09 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr12_+_10460417 | 0.09 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr22_+_39101728 | 0.09 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr19_+_36602104 | 0.09 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr3_+_38347427 | 0.09 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr14_-_24615805 | 0.09 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr18_-_72264805 | 0.09 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr19_-_42721819 | 0.09 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr12_-_47219733 | 0.09 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr14_-_77787198 | 0.09 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr3_+_38323785 | 0.09 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr21_+_35445827 | 0.09 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr1_+_18958008 | 0.09 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr10_-_74114714 | 0.09 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr5_+_141016508 | 0.09 |
ENST00000444782.1
ENST00000521367.1 ENST00000297164.3 |
RELL2
|
RELT-like 2 |
| chr6_-_85473073 | 0.09 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr22_-_46644182 | 0.09 |
ENST00000404583.1
ENST00000404744.1 |
CDPF1
|
cysteine-rich, DPF motif domain containing 1 |
| chr1_-_17445930 | 0.09 |
ENST00000375486.4
ENST00000375481.1 ENST00000444885.2 |
PADI2
|
peptidyl arginine deiminase, type II |
| chrX_-_152989798 | 0.09 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr21_+_39644214 | 0.09 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr12_+_2904102 | 0.09 |
ENST00000001008.4
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr6_+_32811885 | 0.09 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr7_+_26591441 | 0.09 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr9_+_6716478 | 0.09 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chrX_-_152989531 | 0.09 |
ENST00000458587.2
ENST00000416815.1 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr9_+_470288 | 0.09 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr11_+_10471836 | 0.09 |
ENST00000444303.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr5_-_172755056 | 0.08 |
ENST00000520648.1
|
STC2
|
stanniocalcin 2 |
| chr7_-_150020750 | 0.08 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr5_+_179078298 | 0.08 |
ENST00000418535.2
ENST00000425471.1 |
AC136604.1
|
Uncharacterized protein |
| chr14_-_24610779 | 0.08 |
ENST00000560403.1
ENST00000419198.2 ENST00000216799.4 |
EMC9
|
ER membrane protein complex subunit 9 |
| chr6_+_89674246 | 0.08 |
ENST00000369474.1
|
AL079342.1
|
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr9_+_33025209 | 0.08 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr17_+_77070978 | 0.08 |
ENST00000539857.2
ENST00000579016.1 ENST00000311595.9 |
ENGASE
|
endo-beta-N-acetylglucosaminidase |
| chr4_-_147442982 | 0.08 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr8_-_38386175 | 0.08 |
ENST00000437935.2
ENST00000358138.1 |
C8orf86
|
chromosome 8 open reading frame 86 |
| chr1_-_161193349 | 0.08 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr8_+_22853345 | 0.08 |
ENST00000522948.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr2_+_88047606 | 0.08 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr6_-_127840048 | 0.08 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr2_+_232826394 | 0.08 |
ENST00000409401.3
ENST00000441279.1 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr20_+_47835884 | 0.08 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr19_-_14629224 | 0.08 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr11_+_65770227 | 0.08 |
ENST00000527348.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr17_-_62208169 | 0.08 |
ENST00000606895.1
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr6_-_131277510 | 0.08 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr16_+_30662085 | 0.08 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr22_-_42526802 | 0.08 |
ENST00000359033.4
ENST00000389970.3 ENST00000360608.5 |
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr17_+_8339340 | 0.08 |
ENST00000580012.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr2_-_139259244 | 0.08 |
ENST00000414911.1
ENST00000431985.1 |
AC097721.2
|
AC097721.2 |
| chr19_+_13842559 | 0.08 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chrX_+_49020121 | 0.08 |
ENST00000415364.1
ENST00000376338.3 ENST00000425285.1 |
MAGIX
|
MAGI family member, X-linked |
| chr3_-_51533966 | 0.08 |
ENST00000504652.1
|
VPRBP
|
Vpr (HIV-1) binding protein |
| chr1_-_79472365 | 0.08 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr17_+_5323492 | 0.08 |
ENST00000405578.4
ENST00000574003.1 |
RPAIN
|
RPA interacting protein |
| chr2_-_9563216 | 0.08 |
ENST00000467606.1
ENST00000494563.1 ENST00000460001.1 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr7_+_150065278 | 0.08 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr11_+_61197572 | 0.07 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr19_-_1132207 | 0.07 |
ENST00000438103.2
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr17_+_4901199 | 0.07 |
ENST00000320785.5
ENST00000574165.1 |
KIF1C
|
kinesin family member 1C |
| chr12_+_6898638 | 0.07 |
ENST00000011653.4
|
CD4
|
CD4 molecule |
| chr9_-_130667592 | 0.07 |
ENST00000447681.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr12_+_50794891 | 0.07 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr6_+_31465849 | 0.07 |
ENST00000399150.3
|
MICB
|
MHC class I polypeptide-related sequence B |
| chr11_-_47870019 | 0.07 |
ENST00000378460.2
|
NUP160
|
nucleoporin 160kDa |
| chr7_-_102715263 | 0.07 |
ENST00000379305.3
|
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chr7_+_129007964 | 0.07 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr1_+_21877753 | 0.07 |
ENST00000374832.1
|
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr4_-_77819002 | 0.07 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr10_+_24528108 | 0.07 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr19_-_55660561 | 0.07 |
ENST00000587758.1
ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr13_-_79979919 | 0.07 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr19_+_45349630 | 0.07 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr17_+_65027509 | 0.07 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr11_+_94706973 | 0.07 |
ENST00000536741.1
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr6_+_10521574 | 0.07 |
ENST00000495262.1
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr18_-_70931689 | 0.07 |
ENST00000581862.1
|
RP11-169F17.1
|
Protein LOC400655 |
| chr1_+_67773044 | 0.07 |
ENST00000262345.1
ENST00000371000.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.5 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.6 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.4 | GO:0040010 | positive regulation of growth rate(GO:0040010) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.5 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.2 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.6 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.0 | GO:1904000 | positive regulation of eating behavior(GO:1904000) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:0036482 | enzyme active site formation(GO:0018307) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.4 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.1 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.0 | 0.1 | GO:0035698 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) T cell extravasation(GO:0072683) regulation of T cell extravasation(GO:2000407) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.0 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:1990770 | regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) small intestine smooth muscle contraction(GO:1990770) |
| 0.0 | 0.1 | GO:0090345 | alkaloid catabolic process(GO:0009822) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.0 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.3 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:1903487 | epithelial cell differentiation involved in salivary gland development(GO:0060690) epithelial cell maturation involved in salivary gland development(GO:0060691) regulation of plasma cell differentiation(GO:1900098) positive regulation of plasma cell differentiation(GO:1900100) regulation of lactation(GO:1903487) positive regulation of lactation(GO:1903489) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.1 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.0 | 0.2 | GO:0051001 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.0 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.0 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.4 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.2 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |