Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
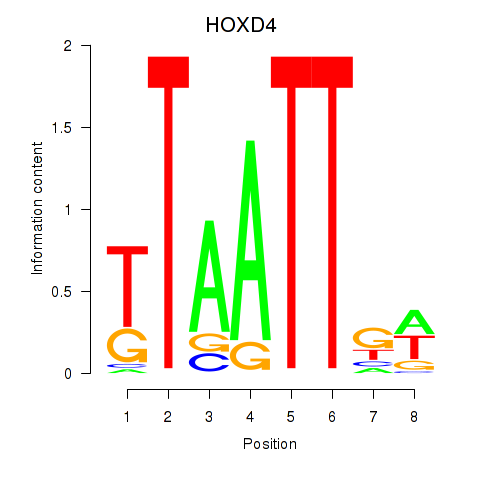
Results for HOXD4
Z-value: 0.33
Transcription factors associated with HOXD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD4
|
ENSG00000170166.5 | homeobox D4 |
Activity profile of HOXD4 motif
Sorted Z-values of HOXD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_97562440 | 0.19 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr4_-_103940791 | 0.15 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr5_+_102200948 | 0.14 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_195310802 | 0.11 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr16_-_21663950 | 0.11 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr6_+_46761118 | 0.10 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr5_-_54988559 | 0.09 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chrX_+_37865804 | 0.08 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr6_+_130339710 | 0.08 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr2_+_190541153 | 0.07 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr5_+_111755280 | 0.07 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr2_-_203735484 | 0.07 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr13_-_54706954 | 0.07 |
ENST00000606706.1
ENST00000607494.1 ENST00000427299.2 ENST00000423442.2 ENST00000451744.1 |
LINC00458
|
long intergenic non-protein coding RNA 458 |
| chr11_-_77790865 | 0.07 |
ENST00000534029.1
ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2
NDUFC2-KCTD14
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr13_+_51913819 | 0.07 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr12_-_118628315 | 0.07 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr5_-_39274617 | 0.06 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chrX_+_56590002 | 0.06 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr3_+_177159695 | 0.06 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr5_+_67586465 | 0.06 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_-_26250835 | 0.06 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr5_+_59783941 | 0.06 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr2_-_203735586 | 0.06 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr4_-_185694872 | 0.06 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr1_-_101360205 | 0.05 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr11_-_104972158 | 0.05 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr8_-_102803163 | 0.05 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr2_-_207078086 | 0.05 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr5_+_95066823 | 0.05 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr4_-_105416039 | 0.05 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr4_+_183065793 | 0.05 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr11_-_64684672 | 0.05 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr18_-_19283649 | 0.05 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr8_-_18871159 | 0.05 |
ENST00000327040.8
ENST00000440756.2 |
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr13_+_37581115 | 0.05 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr3_+_140981456 | 0.05 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr11_-_89224299 | 0.05 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr19_+_50016610 | 0.04 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr1_-_101360374 | 0.04 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr8_-_72274467 | 0.04 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr13_+_110958124 | 0.04 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chrX_-_92928557 | 0.04 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr8_+_42873548 | 0.04 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr1_+_159750776 | 0.04 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr11_-_16419067 | 0.04 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_175462456 | 0.04 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr1_-_101360331 | 0.04 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr3_-_178103144 | 0.04 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr4_-_186570679 | 0.04 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr15_+_84115868 | 0.04 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr2_-_228497888 | 0.04 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chrX_+_139791917 | 0.04 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr3_+_97483572 | 0.04 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr8_-_116504448 | 0.04 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_-_99279928 | 0.04 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_-_207078154 | 0.04 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr2_-_159237472 | 0.04 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr7_+_77428066 | 0.04 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr12_+_9066472 | 0.04 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr18_-_74839891 | 0.04 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr7_+_77428149 | 0.04 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr5_+_68860949 | 0.04 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr16_-_29910853 | 0.04 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr18_-_21891460 | 0.03 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr18_+_29027696 | 0.03 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr3_+_142342240 | 0.03 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr13_+_33160553 | 0.03 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr19_-_5903714 | 0.03 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr19_+_48828582 | 0.03 |
ENST00000270221.6
ENST00000596315.1 |
EMP3
|
epithelial membrane protein 3 |
| chr1_+_84630645 | 0.03 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_+_95848824 | 0.03 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr1_-_151762900 | 0.03 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr2_+_32502952 | 0.03 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr15_-_50558223 | 0.03 |
ENST00000267845.3
|
HDC
|
histidine decarboxylase |
| chr5_+_157158205 | 0.03 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr20_+_18447771 | 0.03 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr8_-_124279627 | 0.03 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr6_-_26108355 | 0.03 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr12_+_28605426 | 0.03 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr11_-_89224638 | 0.03 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr11_+_101983176 | 0.03 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr9_+_130026756 | 0.02 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr7_-_77427676 | 0.02 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr3_-_149293990 | 0.02 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr4_+_130014836 | 0.02 |
ENST00000502887.1
|
C4orf33
|
chromosome 4 open reading frame 33 |
| chr1_-_231560790 | 0.02 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr2_+_166095898 | 0.02 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr2_+_171034646 | 0.02 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr1_-_92952433 | 0.02 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr14_+_23067146 | 0.02 |
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4 |
| chr1_-_151762943 | 0.02 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr1_-_9953295 | 0.02 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr7_+_120629653 | 0.02 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr22_+_44427230 | 0.02 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr17_-_26733179 | 0.02 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr14_+_23067166 | 0.02 |
ENST00000216327.6
ENST00000542041.1 |
ABHD4
|
abhydrolase domain containing 4 |
| chr5_-_111754948 | 0.02 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr10_-_52008313 | 0.02 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr16_+_87636474 | 0.02 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr15_-_54025300 | 0.02 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr3_-_18480260 | 0.02 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr2_+_109237717 | 0.02 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr1_+_202385953 | 0.02 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_-_14372870 | 0.02 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr7_+_55433131 | 0.02 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr1_+_209602771 | 0.02 |
ENST00000440276.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr16_+_24549014 | 0.02 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr1_+_117963209 | 0.02 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr8_+_79503458 | 0.02 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr13_+_20207782 | 0.02 |
ENST00000414242.2
ENST00000361479.5 |
MPHOSPH8
|
M-phase phosphoprotein 8 |
| chr11_-_36310958 | 0.02 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr11_-_72492878 | 0.02 |
ENST00000535054.1
ENST00000545082.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr17_-_73844722 | 0.02 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chr20_+_32150140 | 0.01 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr6_-_36515177 | 0.01 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr1_+_149754227 | 0.01 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr6_+_53659746 | 0.01 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chrX_+_77166172 | 0.01 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr8_+_107738343 | 0.01 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr18_-_51750948 | 0.01 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr1_+_42846443 | 0.01 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr4_-_85771168 | 0.01 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr19_-_14064114 | 0.01 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr2_-_190927447 | 0.01 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr5_-_54988448 | 0.01 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr2_+_187371440 | 0.01 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr3_-_124774802 | 0.01 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr1_+_186798073 | 0.01 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr3_-_126327398 | 0.01 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr15_+_64386261 | 0.01 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr5_+_140762268 | 0.01 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chrX_-_10645773 | 0.01 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr5_+_140743859 | 0.01 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr11_-_89224508 | 0.01 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr2_-_160472952 | 0.01 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr12_+_32654965 | 0.01 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_-_15548591 | 0.01 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr2_+_158114051 | 0.01 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr12_+_12938541 | 0.01 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr7_-_104909435 | 0.01 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr13_-_61989655 | 0.01 |
ENST00000409204.4
|
PCDH20
|
protocadherin 20 |
| chr2_+_191045656 | 0.01 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr4_-_130014705 | 0.01 |
ENST00000503401.1
|
SCLT1
|
sodium channel and clathrin linker 1 |
| chr14_-_24911971 | 0.01 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr15_-_55700457 | 0.01 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr11_-_89224139 | 0.01 |
ENST00000413594.2
|
NOX4
|
NADPH oxidase 4 |
| chr8_+_28196157 | 0.01 |
ENST00000522209.1
|
PNOC
|
prepronociceptin |
| chr20_-_43729750 | 0.01 |
ENST00000537075.1
ENST00000306117.1 |
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr11_-_89224488 | 0.01 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr19_+_30433110 | 0.01 |
ENST00000542441.2
ENST00000392271.1 |
URI1
|
URI1, prefoldin-like chaperone |
| chr15_+_80364901 | 0.01 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr1_-_234831899 | 0.01 |
ENST00000442382.1
|
RP4-781K5.6
|
RP4-781K5.6 |
| chrX_+_149867681 | 0.01 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr9_+_273038 | 0.01 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr1_+_214161854 | 0.01 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr11_-_89223883 | 0.01 |
ENST00000528341.1
|
NOX4
|
NADPH oxidase 4 |
| chr2_+_234590556 | 0.01 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr19_+_48828788 | 0.01 |
ENST00000594198.1
ENST00000597279.1 ENST00000593437.1 |
EMP3
|
epithelial membrane protein 3 |
| chr3_-_4508925 | 0.01 |
ENST00000534863.1
ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1
|
sulfatase modifying factor 1 |
| chr14_+_61449076 | 0.01 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr14_-_101036119 | 0.01 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr2_-_166060571 | 0.01 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr1_+_150039787 | 0.01 |
ENST00000535106.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr10_+_13652047 | 0.01 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr5_+_52083730 | 0.01 |
ENST00000282588.6
ENST00000274311.2 |
ITGA1
PELO
|
integrin, alpha 1 pelota homolog (Drosophila) |
| chr3_+_15045419 | 0.01 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr6_-_44281043 | 0.01 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr6_-_49712147 | 0.01 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr16_-_15736953 | 0.01 |
ENST00000548025.1
ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430
|
KIAA0430 |
| chr9_+_470288 | 0.01 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr2_-_160473114 | 0.01 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr4_-_39979576 | 0.01 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr17_-_29624343 | 0.01 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr10_+_18629628 | 0.00 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_-_152149033 | 0.00 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr17_+_68164752 | 0.00 |
ENST00000535240.1
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr20_-_45980621 | 0.00 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr11_-_102714534 | 0.00 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr22_-_29107919 | 0.00 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr15_-_64385981 | 0.00 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr17_+_66521936 | 0.00 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_-_238108575 | 0.00 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr12_-_81763184 | 0.00 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr2_-_176046391 | 0.00 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr15_-_65809991 | 0.00 |
ENST00000559526.1
ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8
|
dipeptidyl-peptidase 8 |
| chr9_-_107361788 | 0.00 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr2_+_170440844 | 0.00 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr2_-_208030295 | 0.00 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_+_63638339 | 0.00 |
ENST00000343837.3
ENST00000469440.1 |
SNTN
|
sentan, cilia apical structure protein |
| chr7_+_29186192 | 0.00 |
ENST00000539406.1
|
CHN2
|
chimerin 2 |
| chr2_+_110656005 | 0.00 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr18_+_22040620 | 0.00 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr10_+_86184676 | 0.00 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr20_+_5931497 | 0.00 |
ENST00000378886.2
ENST00000265187.4 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr3_-_18480173 | 0.00 |
ENST00000414509.1
|
SATB1
|
SATB homeobox 1 |
| chr1_-_28384598 | 0.00 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chrX_-_13835147 | 0.00 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr4_-_46996424 | 0.00 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr2_-_166060552 | 0.00 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr12_-_112279694 | 0.00 |
ENST00000443596.1
ENST00000442119.1 |
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.0 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |