Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
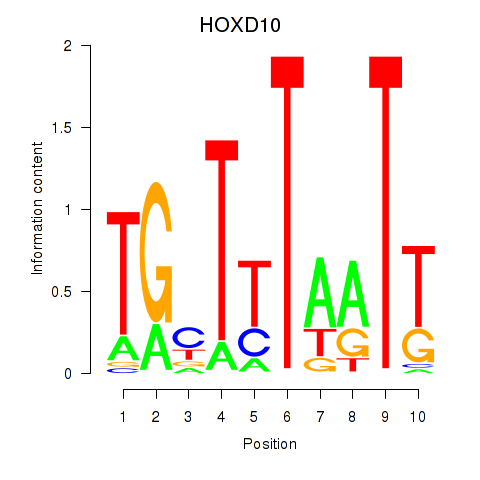
Results for HOXD10
Z-value: 0.41
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.5 | homeobox D10 |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_10601963 | 0.56 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr2_+_90458201 | 0.44 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chrX_-_30871004 | 0.35 |
ENST00000378928.1
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr1_+_223354486 | 0.26 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr17_+_48823896 | 0.21 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr2_+_179318295 | 0.21 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr12_+_20963632 | 0.18 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr9_-_75488984 | 0.17 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr12_-_118796971 | 0.16 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr2_-_188312971 | 0.16 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr15_-_33180439 | 0.16 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr22_-_38577782 | 0.15 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr13_+_100258907 | 0.14 |
ENST00000376355.3
ENST00000376360.1 ENST00000444838.2 ENST00000376354.1 ENST00000339105.4 |
CLYBL
|
citrate lyase beta like |
| chr9_+_42671887 | 0.14 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr2_+_120436760 | 0.14 |
ENST00000445518.1
ENST00000409951.1 |
TMEM177
|
transmembrane protein 177 |
| chr1_+_62439037 | 0.14 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr4_-_76944621 | 0.14 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr4_-_23735183 | 0.12 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr15_+_76352178 | 0.12 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr3_-_126327398 | 0.12 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr1_+_164600184 | 0.12 |
ENST00000482110.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr7_+_116595028 | 0.12 |
ENST00000397751.1
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr3_-_100551141 | 0.12 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr2_+_207804278 | 0.11 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr6_-_32498046 | 0.11 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr12_-_10022735 | 0.11 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr7_+_93652116 | 0.10 |
ENST00000415536.1
|
AC003092.1
|
AC003092.1 |
| chr12_-_10324716 | 0.10 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chrX_-_102565858 | 0.10 |
ENST00000449185.1
ENST00000536889.1 |
BEX2
|
brain expressed X-linked 2 |
| chr1_+_170501270 | 0.10 |
ENST00000367763.3
ENST00000367762.1 |
GORAB
|
golgin, RAB6-interacting |
| chr5_-_122759032 | 0.10 |
ENST00000510582.3
ENST00000328236.5 ENST00000306481.6 ENST00000508442.2 ENST00000395431.2 |
CEP120
|
centrosomal protein 120kDa |
| chr20_+_52105495 | 0.10 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr2_-_99870744 | 0.10 |
ENST00000409238.1
ENST00000423800.1 |
LYG2
|
lysozyme G-like 2 |
| chr8_+_77318769 | 0.10 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr16_-_14726576 | 0.10 |
ENST00000562896.1
|
PARN
|
poly(A)-specific ribonuclease |
| chr12_+_18891045 | 0.09 |
ENST00000317658.3
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr4_-_100356551 | 0.09 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr22_-_38577731 | 0.09 |
ENST00000335539.3
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr2_+_196313239 | 0.09 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr17_+_19437132 | 0.09 |
ENST00000436810.2
ENST00000270570.4 ENST00000457293.1 ENST00000542886.1 ENST00000575023.1 ENST00000395585.1 |
SLC47A1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr6_-_119031228 | 0.09 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr6_+_160221293 | 0.09 |
ENST00000610273.1
ENST00000392167.3 |
PNLDC1
|
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
| chr8_+_61822605 | 0.09 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr12_-_118796910 | 0.08 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr7_+_95115210 | 0.08 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr12_+_21525818 | 0.08 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr14_+_75988851 | 0.08 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr14_+_76776957 | 0.08 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr21_+_25801041 | 0.08 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr1_+_164528866 | 0.08 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_+_166300084 | 0.08 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr11_-_46638378 | 0.08 |
ENST00000529192.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr1_-_238108575 | 0.08 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr20_-_35274548 | 0.08 |
ENST00000262866.4
|
SLA2
|
Src-like-adaptor 2 |
| chr8_-_112248400 | 0.08 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr6_-_134639180 | 0.08 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_195310802 | 0.07 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr11_+_18433840 | 0.07 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr16_-_30122717 | 0.07 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr2_+_29353520 | 0.07 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr5_-_41794313 | 0.07 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr14_-_36990354 | 0.07 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr8_+_95653840 | 0.07 |
ENST00000520385.1
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr7_-_22233442 | 0.07 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_70725856 | 0.07 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr22_-_29107919 | 0.07 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr3_-_149095652 | 0.07 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr1_+_117963209 | 0.07 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr14_-_58618896 | 0.07 |
ENST00000267485.7
|
C14orf37
|
chromosome 14 open reading frame 37 |
| chr1_+_236558694 | 0.07 |
ENST00000359362.5
|
EDARADD
|
EDAR-associated death domain |
| chr20_-_55934878 | 0.07 |
ENST00000543500.1
|
MTRNR2L3
|
MT-RNR2-like 3 |
| chr4_+_71384257 | 0.06 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr6_-_170101749 | 0.06 |
ENST00000448612.1
|
WDR27
|
WD repeat domain 27 |
| chr6_+_27791862 | 0.06 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr17_+_80843910 | 0.06 |
ENST00000572984.1
|
TBCD
|
tubulin folding cofactor D |
| chr4_-_112993808 | 0.06 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr5_-_58882219 | 0.06 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_+_47630255 | 0.06 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr12_+_47610315 | 0.06 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr18_-_25616519 | 0.06 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chrX_-_109590174 | 0.06 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr3_+_158288999 | 0.06 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr17_+_48610074 | 0.06 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr5_-_180665195 | 0.06 |
ENST00000509148.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr2_-_165424973 | 0.06 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr9_+_71944241 | 0.06 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr3_-_99569821 | 0.06 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr1_-_101360331 | 0.06 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr3_+_158288942 | 0.06 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr11_-_7950209 | 0.06 |
ENST00000309838.2
|
OR10A6
|
olfactory receptor, family 10, subfamily A, member 6 |
| chr5_-_39203093 | 0.05 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr5_+_180467259 | 0.05 |
ENST00000515271.1
|
BTNL9
|
butyrophilin-like 9 |
| chr3_-_161089289 | 0.05 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr3_+_158288960 | 0.05 |
ENST00000484955.1
ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr6_-_130543958 | 0.05 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr4_-_114438763 | 0.05 |
ENST00000509594.1
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr14_-_50506589 | 0.05 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr1_-_101360205 | 0.05 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr21_-_31588365 | 0.05 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr19_-_44388116 | 0.05 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr4_-_89951028 | 0.05 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr8_-_90993869 | 0.05 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr12_-_49333446 | 0.05 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr1_+_87458692 | 0.05 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr17_-_4464081 | 0.05 |
ENST00000574154.1
|
GGT6
|
gamma-glutamyltransferase 6 |
| chr1_+_180199393 | 0.05 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr11_+_58938903 | 0.05 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr1_-_76398077 | 0.05 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr12_-_100656134 | 0.05 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr17_-_67264947 | 0.05 |
ENST00000586811.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr20_-_17539456 | 0.05 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr16_-_71610985 | 0.05 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr14_+_37126765 | 0.05 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr9_-_107367951 | 0.05 |
ENST00000542196.1
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2 |
| chr15_-_93353028 | 0.05 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr6_-_20212630 | 0.05 |
ENST00000324607.7
ENST00000541730.1 ENST00000536798.1 |
MBOAT1
|
membrane bound O-acyltransferase domain containing 1 |
| chr16_-_90096309 | 0.05 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr1_-_149459549 | 0.05 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr15_-_64673665 | 0.05 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr1_+_100598742 | 0.04 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr20_+_4702548 | 0.04 |
ENST00000305817.2
|
PRND
|
prion protein 2 (dublet) |
| chr18_-_10701979 | 0.04 |
ENST00000538948.1
ENST00000285141.4 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr2_-_87248975 | 0.04 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr17_-_4463856 | 0.04 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr14_+_37131058 | 0.04 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr19_-_38183228 | 0.04 |
ENST00000587199.1
|
ZFP30
|
ZFP30 zinc finger protein |
| chr1_+_78383813 | 0.04 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr9_-_107361788 | 0.04 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr14_+_75988768 | 0.04 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr10_-_120925054 | 0.04 |
ENST00000419372.1
ENST00000369131.4 ENST00000330036.6 ENST00000355697.2 |
SFXN4
|
sideroflexin 4 |
| chr7_+_134551583 | 0.04 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chrX_-_102565932 | 0.04 |
ENST00000372674.1
ENST00000372677.3 |
BEX2
|
brain expressed X-linked 2 |
| chr1_+_78470530 | 0.04 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr1_+_52521928 | 0.04 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr12_+_85430110 | 0.04 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr12_+_75784850 | 0.04 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr9_+_4839762 | 0.04 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chrX_-_138724994 | 0.04 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr12_+_14561422 | 0.04 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr1_+_158978768 | 0.04 |
ENST00000447473.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr11_-_126174186 | 0.04 |
ENST00000524964.1
|
RP11-712L6.5
|
Uncharacterized protein |
| chr14_-_23292596 | 0.04 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_-_91546926 | 0.04 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_+_69788576 | 0.04 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr6_-_5261141 | 0.04 |
ENST00000330636.4
ENST00000500576.2 |
LYRM4
|
LYR motif containing 4 |
| chr5_-_111312622 | 0.04 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr3_+_63805017 | 0.04 |
ENST00000295896.8
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr9_-_21482312 | 0.04 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr5_+_39105358 | 0.04 |
ENST00000593965.1
|
AC008964.1
|
AC008964.1 |
| chr13_+_114567131 | 0.04 |
ENST00000608651.1
|
GAS6-AS2
|
GAS6 antisense RNA 2 (head to head) |
| chr4_-_110723194 | 0.04 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr2_-_165698662 | 0.04 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr14_+_31046959 | 0.03 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr1_+_174844645 | 0.03 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr8_+_35092959 | 0.03 |
ENST00000404895.2
|
UNC5D
|
unc-5 homolog D (C. elegans) |
| chr11_+_122753391 | 0.03 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr10_-_90967063 | 0.03 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr5_-_137475071 | 0.03 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr6_+_145118873 | 0.03 |
ENST00000432686.1
ENST00000417142.1 |
UTRN
|
utrophin |
| chr7_+_137761199 | 0.03 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr12_-_13248598 | 0.03 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr7_+_11013491 | 0.03 |
ENST00000403050.3
ENST00000445996.2 |
PHF14
|
PHD finger protein 14 |
| chr18_+_61445205 | 0.03 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_145883868 | 0.03 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr5_+_68513557 | 0.03 |
ENST00000256441.4
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr6_-_8102714 | 0.03 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr13_-_46679185 | 0.03 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr19_-_48389651 | 0.03 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr12_-_118797475 | 0.03 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr18_-_21891460 | 0.03 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr2_+_138722028 | 0.03 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr1_-_101360374 | 0.03 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr5_-_59481406 | 0.03 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_+_210444748 | 0.03 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr17_+_48823975 | 0.03 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr15_-_78538025 | 0.03 |
ENST00000560817.1
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr7_+_5465382 | 0.03 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr1_+_186798073 | 0.03 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chrX_-_19988382 | 0.03 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr2_+_183582774 | 0.03 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr3_+_69811858 | 0.03 |
ENST00000433517.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr11_-_93271058 | 0.03 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chrM_+_10464 | 0.03 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr3_+_89156799 | 0.03 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr18_+_29027696 | 0.03 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr22_+_41258250 | 0.03 |
ENST00000544094.1
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr11_-_26593649 | 0.03 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr15_+_42566384 | 0.03 |
ENST00000440615.2
ENST00000318010.8 |
GANC
|
glucosidase, alpha; neutral C |
| chr6_+_96025341 | 0.03 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr4_+_100495864 | 0.03 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr5_+_140762268 | 0.03 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr6_+_118869452 | 0.03 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr12_+_16109519 | 0.03 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr4_+_76649797 | 0.03 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr12_-_48164812 | 0.03 |
ENST00000549151.1
ENST00000548919.1 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr15_+_41549105 | 0.03 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr6_-_90025011 | 0.03 |
ENST00000402938.3
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr18_+_48918368 | 0.03 |
ENST00000583982.1
ENST00000578152.1 ENST00000583609.1 ENST00000435144.1 ENST00000580841.1 |
RP11-267C16.1
|
RP11-267C16.1 |
| chr10_+_115614370 | 0.03 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr1_-_242612726 | 0.03 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |