Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
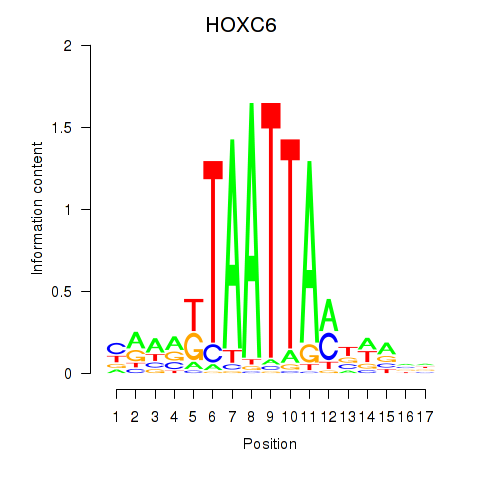
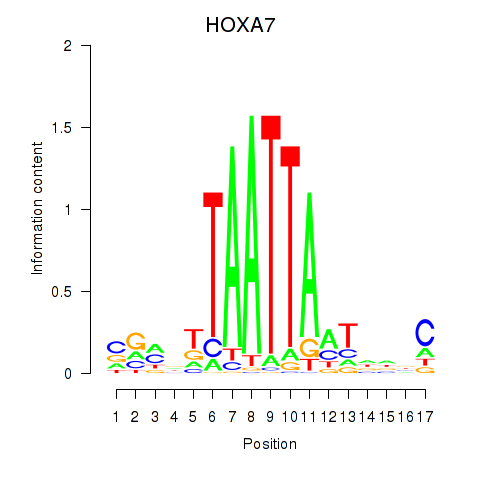
Results for HOXC6_HOXA7
Z-value: 0.21
Transcription factors associated with HOXC6_HOXA7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC6
|
ENSG00000197757.7 | homeobox C6 |
|
HOXA7
|
ENSG00000122592.6 | homeobox A7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA7 | hg19_v2_chr7_-_27196267_27196311 | -0.55 | 2.6e-01 | Click! |
Activity profile of HOXC6_HOXA7 motif
Sorted Z-values of HOXC6_HOXA7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_90248739 | 0.14 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr12_+_78359999 | 0.14 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr12_-_86650077 | 0.07 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr3_+_111718173 | 0.07 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr4_+_147145709 | 0.07 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr7_+_150020363 | 0.06 |
ENST00000359623.4
ENST00000493307.1 |
LRRC61
|
leucine rich repeat containing 61 |
| chr6_-_133035185 | 0.06 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr6_+_46761118 | 0.06 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr7_+_134671234 | 0.06 |
ENST00000436302.2
ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein-like 3 |
| chr7_+_150020329 | 0.06 |
ENST00000323078.7
|
LRRC61
|
leucine rich repeat containing 61 |
| chr17_-_4938712 | 0.05 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr14_+_101295638 | 0.05 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr4_+_71108300 | 0.05 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr12_-_112279694 | 0.05 |
ENST00000443596.1
ENST00000442119.1 |
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1 |
| chr15_+_67418047 | 0.05 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr3_+_111717511 | 0.05 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr1_+_212965170 | 0.05 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr3_+_111717600 | 0.05 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr10_-_27529486 | 0.04 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr3_+_111718036 | 0.04 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr11_-_8857248 | 0.04 |
ENST00000534248.1
ENST00000530959.1 ENST00000531578.1 ENST00000533225.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr14_-_101295407 | 0.04 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr2_+_152214098 | 0.04 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr12_-_94673956 | 0.04 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr5_+_66300464 | 0.04 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_-_54618650 | 0.04 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr2_+_182850551 | 0.04 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_-_151395525 | 0.04 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr1_+_149239529 | 0.04 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr16_+_82068585 | 0.04 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr2_+_228736335 | 0.04 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr17_-_46688334 | 0.04 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr11_-_102651343 | 0.03 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr3_+_138340049 | 0.03 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr19_+_49199209 | 0.03 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr4_+_158493642 | 0.03 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr11_-_63376013 | 0.03 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr13_-_30880979 | 0.03 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr10_+_86184676 | 0.03 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr6_-_47445214 | 0.03 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr9_+_125133467 | 0.03 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr15_-_55657428 | 0.03 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr2_+_105050794 | 0.03 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chrX_+_84258832 | 0.03 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr1_-_92952433 | 0.03 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_-_92777606 | 0.03 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr2_+_182850743 | 0.03 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chrX_+_10126488 | 0.03 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr12_+_56435637 | 0.03 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr15_+_63188009 | 0.03 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr8_-_133772870 | 0.03 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr9_-_5339873 | 0.02 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr3_-_141719195 | 0.02 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr8_+_27238147 | 0.02 |
ENST00000412793.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chrX_-_110655306 | 0.02 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr16_+_15489629 | 0.02 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr15_+_64680003 | 0.02 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr2_+_149974684 | 0.02 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr5_+_66300446 | 0.02 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_-_91573132 | 0.02 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr18_+_32558208 | 0.02 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_-_176046391 | 0.02 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr19_-_54619006 | 0.02 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr13_-_81801115 | 0.02 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr3_-_54962100 | 0.02 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr4_-_119759795 | 0.02 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chrX_-_14047996 | 0.02 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr11_+_126173647 | 0.02 |
ENST00000263579.4
|
DCPS
|
decapping enzyme, scavenger |
| chr10_-_99205607 | 0.02 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr7_-_22862448 | 0.02 |
ENST00000358435.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr19_+_12175535 | 0.02 |
ENST00000550826.1
|
ZNF844
|
zinc finger protein 844 |
| chr6_+_130339710 | 0.02 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr5_-_16916624 | 0.02 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr21_-_35796241 | 0.02 |
ENST00000450895.1
|
AP000322.53
|
AP000322.53 |
| chr12_-_91573249 | 0.02 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr19_+_55043977 | 0.02 |
ENST00000335056.3
|
KIR3DX1
|
killer cell immunoglobulin-like receptor, three domains, X1 |
| chr18_-_47018869 | 0.02 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr2_-_128284020 | 0.02 |
ENST00000295321.4
ENST00000455721.2 |
IWS1
|
IWS1 homolog (S. cerevisiae) |
| chr11_-_111649074 | 0.02 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr7_-_150020578 | 0.02 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr1_-_201140673 | 0.01 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr17_+_35851570 | 0.01 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr8_-_133772794 | 0.01 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr7_-_107968999 | 0.01 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr4_-_68829226 | 0.01 |
ENST00000396188.2
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr2_+_25015968 | 0.01 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr11_+_62496114 | 0.01 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr3_+_45927994 | 0.01 |
ENST00000357632.2
ENST00000395963.2 |
CCR9
|
chemokine (C-C motif) receptor 9 |
| chrX_+_43515467 | 0.01 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr12_-_91573316 | 0.01 |
ENST00000393155.1
|
DCN
|
decorin |
| chr2_+_202098203 | 0.01 |
ENST00000450491.1
ENST00000440732.1 ENST00000392258.3 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr2_+_27498289 | 0.01 |
ENST00000296097.3
ENST00000420191.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr16_+_82068873 | 0.01 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr1_-_232651312 | 0.01 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr2_-_40680578 | 0.01 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr10_-_101825151 | 0.01 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr7_-_44122063 | 0.01 |
ENST00000335195.6
ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM
|
polymerase (DNA directed), mu |
| chr8_-_93115445 | 0.01 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_-_130080977 | 0.01 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr17_+_72931876 | 0.01 |
ENST00000328801.4
|
OTOP3
|
otopetrin 3 |
| chr1_+_15668240 | 0.01 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr8_+_70476088 | 0.01 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chr11_+_2405833 | 0.01 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr6_+_44355257 | 0.01 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr4_-_68829144 | 0.01 |
ENST00000508048.1
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr7_+_134528635 | 0.01 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr15_+_25068773 | 0.01 |
ENST00000400100.1
ENST00000400098.1 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr2_-_25391425 | 0.01 |
ENST00000264708.3
ENST00000449220.1 |
POMC
|
proopiomelanocortin |
| chr2_-_25391468 | 0.01 |
ENST00000395826.2
|
POMC
|
proopiomelanocortin |
| chr8_+_77593448 | 0.01 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr7_-_105029812 | 0.01 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr1_-_186430222 | 0.01 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr14_-_23426231 | 0.01 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr10_+_18549645 | 0.01 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr12_+_26348246 | 0.01 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr16_-_29934558 | 0.01 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr1_+_66820058 | 0.01 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr5_+_89770696 | 0.01 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr21_-_43346790 | 0.01 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr6_+_30029008 | 0.01 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr15_+_89922345 | 0.01 |
ENST00000558982.1
|
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr12_+_112279782 | 0.01 |
ENST00000550735.2
|
MAPKAPK5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr19_-_5903714 | 0.01 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr12_+_26348429 | 0.01 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr6_+_26365387 | 0.01 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr14_+_56584414 | 0.01 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chrX_-_77225135 | 0.01 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr6_+_26402517 | 0.01 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr5_+_89770664 | 0.01 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr2_-_74619152 | 0.01 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr11_-_104827425 | 0.01 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_-_107729887 | 0.01 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_-_138453606 | 0.01 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr10_-_112678692 | 0.01 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr5_-_22853429 | 0.01 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr12_+_131438443 | 0.01 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr1_+_8021954 | 0.01 |
ENST00000377491.1
ENST00000377488.1 |
PARK7
|
parkinson protein 7 |
| chr11_-_77790865 | 0.01 |
ENST00000534029.1
ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2
NDUFC2-KCTD14
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr12_+_107078474 | 0.01 |
ENST00000552866.1
ENST00000229387.5 |
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr14_-_23058063 | 0.01 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.4 |
DAD1
|
defender against cell death 1 |
| chr18_-_74839891 | 0.01 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr11_-_31391276 | 0.01 |
ENST00000452803.1
|
DCDC1
|
doublecortin domain containing 1 |
| chrM_+_10053 | 0.01 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr7_-_44580861 | 0.01 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr4_-_152149033 | 0.01 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr6_-_138833630 | 0.01 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr11_-_77791156 | 0.01 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr11_+_89764274 | 0.01 |
ENST00000448984.1
ENST00000432771.1 |
TRIM49C
|
tripartite motif containing 49C |
| chr11_+_33061543 | 0.01 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr17_-_76220740 | 0.01 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr1_-_160924589 | 0.01 |
ENST00000368029.3
|
ITLN2
|
intelectin 2 |
| chr2_+_234826016 | 0.01 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr17_-_72772462 | 0.01 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_-_128894053 | 0.00 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr19_-_36001113 | 0.00 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr17_-_39623681 | 0.00 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr1_-_212965104 | 0.00 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr2_-_74618964 | 0.00 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr17_+_61151306 | 0.00 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr9_-_215744 | 0.00 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr10_-_45811056 | 0.00 |
ENST00000553795.1
|
OR13A1
|
olfactory receptor, family 13, subfamily A, member 1 |
| chr18_-_47018769 | 0.00 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr6_-_138866823 | 0.00 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr4_+_169418255 | 0.00 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_170632250 | 0.00 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr2_-_25391507 | 0.00 |
ENST00000380794.1
|
POMC
|
proopiomelanocortin |
| chr18_+_32558380 | 0.00 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_+_202047596 | 0.00 |
ENST00000286186.6
ENST00000360132.3 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr7_-_32338917 | 0.00 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr7_-_55620433 | 0.00 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr11_+_112041253 | 0.00 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chrM_+_10464 | 0.00 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr1_+_221051699 | 0.00 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr12_-_89919765 | 0.00 |
ENST00000541909.1
ENST00000313546.3 |
POC1B
|
POC1 centriolar protein B |
| chr16_+_53469525 | 0.00 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr2_+_202098166 | 0.00 |
ENST00000392263.2
ENST00000264274.9 ENST00000392259.2 ENST00000392266.3 ENST00000432109.2 ENST00000264275.5 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr3_+_157154578 | 0.00 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr11_-_111649015 | 0.00 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_+_2933893 | 0.00 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr2_+_202047843 | 0.00 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr5_+_55149150 | 0.00 |
ENST00000297015.3
|
IL31RA
|
interleukin 31 receptor A |
| chr4_-_100009856 | 0.00 |
ENST00000296412.8
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr2_-_25391772 | 0.00 |
ENST00000405623.1
|
POMC
|
proopiomelanocortin |
| chr2_-_74618907 | 0.00 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr19_+_12175596 | 0.00 |
ENST00000441304.2
|
ZNF844
|
zinc finger protein 844 |
| chr5_+_162930114 | 0.00 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr11_-_89541743 | 0.00 |
ENST00000329758.1
|
TRIM49
|
tripartite motif containing 49 |
| chr12_-_52761262 | 0.00 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr18_-_47018897 | 0.00 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr1_-_38019878 | 0.00 |
ENST00000296215.6
|
SNIP1
|
Smad nuclear interacting protein 1 |
| chr13_-_30881134 | 0.00 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr6_+_26402465 | 0.00 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr6_-_34639733 | 0.00 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr2_+_26624775 | 0.00 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr6_+_149721495 | 0.00 |
ENST00000326669.4
|
SUMO4
|
small ubiquitin-like modifier 4 |
| chr17_-_72772425 | 0.00 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_-_26593677 | 0.00 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr12_+_79371565 | 0.00 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr5_-_20575959 | 0.00 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr4_+_169418195 | 0.00 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC6_HOXA7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |