Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
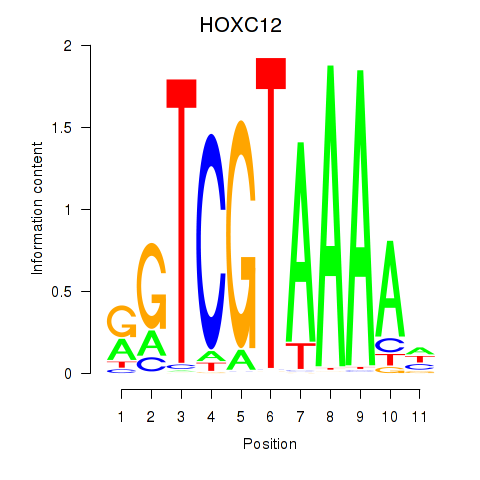
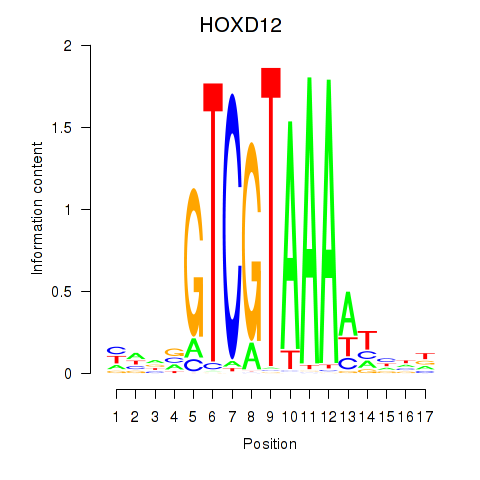
Results for HOXC12_HOXD12
Z-value: 0.67
Transcription factors associated with HOXC12_HOXD12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC12
|
ENSG00000123407.3 | homeobox C12 |
|
HOXD12
|
ENSG00000170178.5 | homeobox D12 |
Activity profile of HOXC12_HOXD12 motif
Sorted Z-values of HOXC12_HOXD12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_73455788 | 0.64 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr12_+_113354341 | 0.59 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_-_153113927 | 0.48 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr7_+_76101379 | 0.44 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr14_-_50065882 | 0.44 |
ENST00000539688.1
|
AL139099.1
|
Full-length cDNA clone CS0DK012YO09 of HeLa cells of Homo sapiens (human); Uncharacterized protein |
| chr15_+_41245160 | 0.39 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr11_-_104480019 | 0.38 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr19_+_7745708 | 0.35 |
ENST00000596148.1
ENST00000317378.5 ENST00000426877.2 |
TRAPPC5
|
trafficking protein particle complex 5 |
| chr18_+_3466248 | 0.35 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr11_-_9336117 | 0.30 |
ENST00000527813.1
ENST00000533723.1 |
TMEM41B
|
transmembrane protein 41B |
| chr17_-_7760457 | 0.29 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr19_-_55677999 | 0.28 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr5_+_159656437 | 0.28 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr1_-_155948890 | 0.27 |
ENST00000471589.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr22_-_20307532 | 0.27 |
ENST00000405465.3
ENST00000248879.3 |
DGCR6L
|
DiGeorge syndrome critical region gene 6-like |
| chr10_+_118608998 | 0.25 |
ENST00000409522.1
ENST00000341276.5 ENST00000512864.2 |
ENO4
|
enolase family member 4 |
| chr22_+_22676808 | 0.25 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr4_-_69536346 | 0.23 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr19_+_13885252 | 0.23 |
ENST00000221576.4
|
C19orf53
|
chromosome 19 open reading frame 53 |
| chr7_-_41742697 | 0.20 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr7_-_27179814 | 0.20 |
ENST00000522788.1
ENST00000521779.1 |
HOXA3
|
homeobox A3 |
| chr2_+_149974684 | 0.19 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr16_+_2273558 | 0.18 |
ENST00000301727.4
ENST00000565090.1 ENST00000564139.1 |
E4F1
|
E4F transcription factor 1 |
| chr16_+_2273645 | 0.18 |
ENST00000565413.1
|
E4F1
|
E4F transcription factor 1 |
| chr5_+_35852797 | 0.17 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr3_-_57326704 | 0.17 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr15_+_66797627 | 0.17 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr1_-_11986442 | 0.17 |
ENST00000376572.3
ENST00000376576.3 |
KIAA2013
|
KIAA2013 |
| chr6_-_26199471 | 0.17 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr6_-_26199499 | 0.17 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr1_-_146989697 | 0.17 |
ENST00000437831.1
|
LINC00624
|
long intergenic non-protein coding RNA 624 |
| chr1_+_15736359 | 0.16 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr7_+_76090993 | 0.16 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr7_+_138915102 | 0.16 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr8_+_121925301 | 0.16 |
ENST00000517739.1
|
RP11-369K17.1
|
RP11-369K17.1 |
| chr1_+_152784447 | 0.16 |
ENST00000360090.3
|
LCE1B
|
late cornified envelope 1B |
| chr2_+_131769256 | 0.15 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_-_128049955 | 0.15 |
ENST00000419067.2
ENST00000378717.4 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr17_-_42767092 | 0.15 |
ENST00000588687.1
|
CCDC43
|
coiled-coil domain containing 43 |
| chr1_-_247242048 | 0.15 |
ENST00000366503.2
|
ZNF670
|
zinc finger protein 670 |
| chr2_-_113810444 | 0.15 |
ENST00000259213.4
ENST00000327407.2 |
IL36B
|
interleukin 36, beta |
| chr19_-_48759119 | 0.14 |
ENST00000522889.1
ENST00000520753.1 ENST00000519940.1 ENST00000519332.1 ENST00000521437.1 ENST00000520007.1 ENST00000521613.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr22_+_38035623 | 0.14 |
ENST00000336738.5
ENST00000442465.2 |
SH3BP1
|
SH3-domain binding protein 1 |
| chr11_-_102668879 | 0.14 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr7_-_76955563 | 0.14 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chr13_+_113030658 | 0.14 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr22_+_18893816 | 0.14 |
ENST00000608842.1
|
DGCR6
|
DiGeorge syndrome critical region gene 6 |
| chr15_-_99789736 | 0.14 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr19_-_11457162 | 0.14 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr19_-_48753028 | 0.14 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr11_-_128775930 | 0.14 |
ENST00000524878.1
|
C11orf45
|
chromosome 11 open reading frame 45 |
| chr8_-_144699628 | 0.14 |
ENST00000529048.1
ENST00000529064.1 |
TSTA3
|
tissue specific transplantation antigen P35B |
| chr17_+_7517264 | 0.13 |
ENST00000593717.1
ENST00000572182.1 ENST00000574539.1 ENST00000576728.1 ENST00000575314.1 ENST00000570547.1 ENST00000572262.1 ENST00000576478.1 |
AC007421.1
SHBG
|
AC007421.1 sex hormone-binding globulin |
| chr6_-_112081113 | 0.13 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr10_+_51549498 | 0.13 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr8_-_144699668 | 0.13 |
ENST00000425753.2
|
TSTA3
|
tissue specific transplantation antigen P35B |
| chr11_-_85376121 | 0.13 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr11_-_7904464 | 0.13 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr2_-_27435390 | 0.13 |
ENST00000428518.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr14_+_39944025 | 0.13 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr2_+_37571717 | 0.13 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr17_+_6679528 | 0.13 |
ENST00000321535.4
|
FBXO39
|
F-box protein 39 |
| chr1_+_171060018 | 0.13 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr19_-_5680202 | 0.12 |
ENST00000590389.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr3_+_75713481 | 0.12 |
ENST00000308062.3
ENST00000464571.1 |
FRG2C
|
FSHD region gene 2 family, member C |
| chr4_-_682960 | 0.12 |
ENST00000512249.1
ENST00000515118.1 ENST00000347950.5 |
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr11_+_67351213 | 0.12 |
ENST00000398603.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr12_-_31743901 | 0.12 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr1_+_92545862 | 0.12 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr11_+_46366918 | 0.12 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr16_+_67906919 | 0.12 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr19_+_49496782 | 0.12 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr19_-_12267524 | 0.12 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr19_-_11457082 | 0.11 |
ENST00000587948.1
|
TMEM205
|
transmembrane protein 205 |
| chr17_-_29624343 | 0.11 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr17_+_17685422 | 0.11 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr11_+_46366799 | 0.11 |
ENST00000532868.2
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr6_-_31681839 | 0.11 |
ENST00000409239.1
ENST00000461287.1 |
LY6G6E
XXbac-BPG32J3.20
|
lymphocyte antigen 6 complex, locus G6E (pseudogene) Uncharacterized protein |
| chr1_+_219347203 | 0.11 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr19_-_11456872 | 0.11 |
ENST00000586218.1
|
TMEM205
|
transmembrane protein 205 |
| chr9_-_96108696 | 0.11 |
ENST00000375419.1
|
C9orf129
|
chromosome 9 open reading frame 129 |
| chr15_+_101459420 | 0.11 |
ENST00000388948.3
ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1
|
leucine-rich repeat kinase 1 |
| chr6_-_127840048 | 0.11 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr11_+_89764274 | 0.11 |
ENST00000448984.1
ENST00000432771.1 |
TRIM49C
|
tripartite motif containing 49C |
| chrX_+_153775821 | 0.10 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr15_-_79576229 | 0.10 |
ENST00000557790.1
ENST00000558297.1 |
RP11-17L5.4
|
RP11-17L5.4 |
| chr6_+_13272904 | 0.10 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr11_+_63606373 | 0.10 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr8_-_42065187 | 0.10 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chr11_+_61891445 | 0.10 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr10_+_12237924 | 0.10 |
ENST00000429258.2
ENST00000281141.4 |
CDC123
|
cell division cycle 123 |
| chr11_-_796197 | 0.10 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr6_-_169652246 | 0.10 |
ENST00000435791.1
|
THBS2
|
thrombospondin 2 |
| chr2_-_225362533 | 0.10 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr2_-_70475730 | 0.10 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr18_+_61223393 | 0.10 |
ENST00000269491.1
ENST00000382768.1 |
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr7_+_150065278 | 0.10 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr19_-_55677920 | 0.10 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr7_-_78400598 | 0.10 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_-_134093738 | 0.10 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr14_-_68000442 | 0.10 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr8_+_118147498 | 0.09 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr2_+_37571845 | 0.09 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr19_+_49496705 | 0.09 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr19_-_11456905 | 0.09 |
ENST00000588560.1
ENST00000592952.1 |
TMEM205
|
transmembrane protein 205 |
| chr1_-_246670614 | 0.09 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr4_+_70894130 | 0.09 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr13_+_50570019 | 0.09 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr8_-_42065075 | 0.09 |
ENST00000429089.2
ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT
|
plasminogen activator, tissue |
| chr19_-_5680231 | 0.09 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr5_+_93954358 | 0.09 |
ENST00000504099.1
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr2_+_172778952 | 0.09 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr1_+_219347186 | 0.09 |
ENST00000366928.5
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr6_+_26199737 | 0.09 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chrX_-_48326764 | 0.09 |
ENST00000413668.1
ENST00000441948.1 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr19_-_49955050 | 0.09 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chrX_+_9935392 | 0.08 |
ENST00000445307.2
|
AC002365.1
|
Homo sapiens uncharacterized LOC100288814 (LOC100288814), mRNA. |
| chr19_-_11456935 | 0.08 |
ENST00000590788.1
ENST00000586590.1 ENST00000589555.1 ENST00000586956.1 ENST00000593256.2 ENST00000447337.1 ENST00000591677.1 ENST00000586701.1 ENST00000589655.1 |
TMEM205
RAB3D
|
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chr14_-_60952739 | 0.08 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr4_-_70626314 | 0.08 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr6_-_27782548 | 0.08 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr1_+_24117627 | 0.08 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr14_+_67291158 | 0.08 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr6_+_108616054 | 0.08 |
ENST00000368977.4
|
LACE1
|
lactation elevated 1 |
| chr16_-_88878305 | 0.08 |
ENST00000569616.1
ENST00000563655.1 ENST00000567713.1 ENST00000426324.2 ENST00000378364.3 |
APRT
|
adenine phosphoribosyltransferase |
| chr1_-_242162375 | 0.08 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr15_+_71184931 | 0.08 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr14_-_77889860 | 0.08 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr7_-_128050027 | 0.08 |
ENST00000343214.4
ENST00000354269.5 ENST00000348127.6 ENST00000497868.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr6_+_108616243 | 0.08 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
| chr3_-_112127981 | 0.08 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr9_-_4299874 | 0.08 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr13_-_47471155 | 0.08 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr1_+_199996702 | 0.08 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr8_-_63998590 | 0.07 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr11_+_67351019 | 0.07 |
ENST00000398606.3
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr1_+_42928945 | 0.07 |
ENST00000428554.2
|
CCDC30
|
coiled-coil domain containing 30 |
| chr22_-_39928823 | 0.07 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr7_+_2443202 | 0.07 |
ENST00000258711.6
|
CHST12
|
carbohydrate (chondroitin 4) sulfotransferase 12 |
| chr19_-_11456722 | 0.07 |
ENST00000354882.5
|
TMEM205
|
transmembrane protein 205 |
| chr10_-_51371321 | 0.07 |
ENST00000602930.1
|
AGAP8
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr19_+_44331493 | 0.07 |
ENST00000588797.1
|
ZNF283
|
zinc finger protein 283 |
| chr6_-_137365402 | 0.07 |
ENST00000541547.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chr17_-_79829190 | 0.07 |
ENST00000581876.1
ENST00000584461.1 ENST00000583868.1 ENST00000400721.4 ENST00000269321.7 |
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr19_+_21106028 | 0.07 |
ENST00000597314.1
ENST00000601924.1 |
ZNF85
|
zinc finger protein 85 |
| chr8_+_22601 | 0.07 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr21_-_46359760 | 0.07 |
ENST00000330551.3
ENST00000397841.1 ENST00000380070.4 |
C21orf67
|
chromosome 21 open reading frame 67 |
| chr14_+_97925151 | 0.07 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr8_+_101170563 | 0.07 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr3_+_179322481 | 0.07 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr5_+_169011033 | 0.07 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_49497121 | 0.06 |
ENST00000413176.2
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr18_+_12703002 | 0.06 |
ENST00000590217.1
|
PSMG2
|
proteasome (prosome, macropain) assembly chaperone 2 |
| chr13_+_78315528 | 0.06 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr5_+_145718587 | 0.06 |
ENST00000230732.4
|
POU4F3
|
POU class 4 homeobox 3 |
| chr13_+_111855399 | 0.06 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr17_-_191188 | 0.06 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr17_-_73127826 | 0.06 |
ENST00000582170.1
ENST00000245552.2 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr17_-_7232585 | 0.06 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr4_-_120243545 | 0.06 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr17_-_66951474 | 0.06 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr6_+_126221034 | 0.06 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr19_-_47987419 | 0.06 |
ENST00000536339.1
ENST00000595554.1 ENST00000600271.1 ENST00000338134.3 |
KPTN
|
kaptin (actin binding protein) |
| chr15_-_72410455 | 0.06 |
ENST00000569314.1
|
MYO9A
|
myosin IXA |
| chr7_+_80275953 | 0.06 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_+_201754050 | 0.06 |
ENST00000426253.1
ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr16_+_20818020 | 0.06 |
ENST00000564274.1
ENST00000563068.1 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr7_-_72992663 | 0.06 |
ENST00000432538.1
|
TBL2
|
transducin (beta)-like 2 |
| chr19_+_18668672 | 0.06 |
ENST00000601630.1
ENST00000599000.1 ENST00000595073.1 ENST00000596785.1 |
KXD1
|
KxDL motif containing 1 |
| chr10_-_70287172 | 0.06 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr19_+_37341752 | 0.06 |
ENST00000586933.1
ENST00000532141.1 ENST00000420450.1 ENST00000526123.1 |
ZNF345
|
zinc finger protein 345 |
| chr15_-_55657428 | 0.06 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr7_-_156433195 | 0.06 |
ENST00000333319.6
|
C7orf13
|
chromosome 7 open reading frame 13 |
| chr3_+_167582561 | 0.06 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr2_-_225811747 | 0.06 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr14_+_38264418 | 0.06 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr19_+_18668616 | 0.06 |
ENST00000600372.1
|
KXD1
|
KxDL motif containing 1 |
| chr19_-_48753064 | 0.06 |
ENST00000520153.1
ENST00000357778.5 ENST00000520015.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr7_-_128050000 | 0.06 |
ENST00000489263.1
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr5_+_140552218 | 0.06 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr4_-_111120334 | 0.06 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr15_-_41836441 | 0.06 |
ENST00000567866.1
ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1
|
RNA polymerase II associated protein 1 |
| chr4_+_56814968 | 0.06 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr11_+_110001723 | 0.06 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr7_+_80275621 | 0.06 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr19_+_11200038 | 0.06 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr21_-_15918618 | 0.05 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr13_-_30160925 | 0.05 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_+_140254884 | 0.05 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr17_-_26127525 | 0.05 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr3_-_19975665 | 0.05 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr14_+_35591020 | 0.05 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr12_+_104850771 | 0.05 |
ENST00000546689.1
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr20_-_44540686 | 0.05 |
ENST00000477313.1
ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP
|
phospholipid transfer protein |
| chr10_+_6625733 | 0.05 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr5_+_140514782 | 0.05 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr6_-_656963 | 0.05 |
ENST00000380907.2
|
HUS1B
|
HUS1 checkpoint homolog b (S. pombe) |
| chr4_+_6202448 | 0.05 |
ENST00000508601.1
|
RP11-586D19.1
|
RP11-586D19.1 |
| chr19_+_2785458 | 0.05 |
ENST00000307741.6
ENST00000585338.1 |
THOP1
|
thimet oligopeptidase 1 |
| chr10_-_106098162 | 0.05 |
ENST00000337478.1
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC12_HOXD12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.2 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 0.3 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.3 | GO:0050577 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |