Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
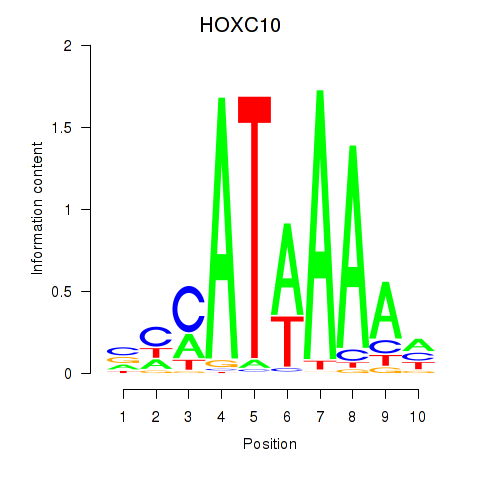
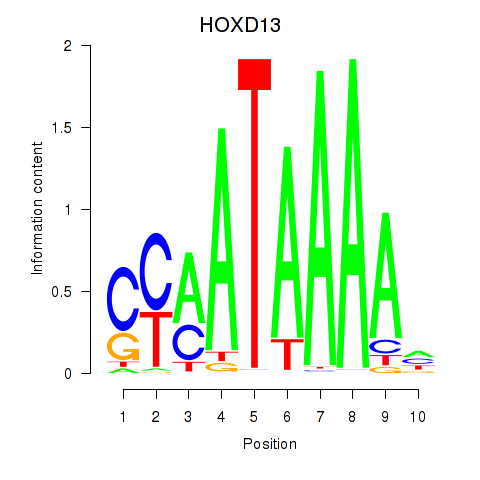
Results for HOXC10_HOXD13
Z-value: 0.53
Transcription factors associated with HOXC10_HOXD13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC10
|
ENSG00000180818.4 | homeobox C10 |
|
HOXD13
|
ENSG00000128714.5 | homeobox D13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC10 | hg19_v2_chr12_+_54378923_54378970 | -0.05 | 9.2e-01 | Click! |
Activity profile of HOXC10_HOXD13 motif
Sorted Z-values of HOXC10_HOXD13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22766766 | 0.90 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr6_+_32812568 | 0.68 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr11_+_93754513 | 0.66 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr7_-_41742697 | 0.64 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr4_+_71384257 | 0.58 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr15_-_80263506 | 0.57 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr6_+_27775899 | 0.53 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr18_+_68002675 | 0.51 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr5_+_147582348 | 0.48 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr6_+_12290586 | 0.41 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr4_+_71384300 | 0.36 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr8_-_73793975 | 0.36 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr11_+_107650219 | 0.32 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr8_-_25281747 | 0.29 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr6_-_27775694 | 0.28 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chrY_+_15418467 | 0.28 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr10_-_106240032 | 0.28 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr1_+_153003671 | 0.28 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr13_-_81801115 | 0.27 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr10_-_69597828 | 0.26 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr18_-_73967160 | 0.25 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr22_-_30642782 | 0.24 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chrX_-_19688475 | 0.22 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr10_-_94301107 | 0.21 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr17_-_38859996 | 0.21 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr1_-_43919346 | 0.20 |
ENST00000372430.3
ENST00000372433.1 ENST00000372434.1 ENST00000486909.1 |
HYI
|
hydroxypyruvate isomerase (putative) |
| chr12_-_52828147 | 0.20 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr21_+_17792672 | 0.19 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_77021702 | 0.18 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_-_46806540 | 0.18 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr11_-_102826434 | 0.17 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr14_-_55369525 | 0.17 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr5_+_147582387 | 0.17 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr22_+_39916558 | 0.17 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr22_+_50981079 | 0.17 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr7_-_77045617 | 0.17 |
ENST00000257626.7
|
GSAP
|
gamma-secretase activating protein |
| chr3_+_173116225 | 0.17 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr11_+_118401706 | 0.16 |
ENST00000411589.2
ENST00000442938.2 ENST00000359862.4 |
TMEM25
|
transmembrane protein 25 |
| chr9_+_130911723 | 0.16 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr12_-_13105081 | 0.15 |
ENST00000541128.1
|
GPRC5D
|
G protein-coupled receptor, family C, group 5, member D |
| chr4_+_169575875 | 0.15 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_+_76745385 | 0.15 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr2_+_138721850 | 0.15 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr14_-_21493123 | 0.15 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr3_+_190105909 | 0.15 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr18_-_5396271 | 0.14 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr14_-_38028689 | 0.14 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chrM_+_4431 | 0.14 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr11_-_132813627 | 0.14 |
ENST00000374778.4
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr3_+_101546827 | 0.14 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr12_+_24857397 | 0.13 |
ENST00000536131.1
|
RP11-625L16.1
|
RP11-625L16.1 |
| chr2_-_216300784 | 0.13 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr11_-_128392085 | 0.13 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr14_+_39703112 | 0.13 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr14_+_104710541 | 0.13 |
ENST00000419115.1
|
C14orf144
|
chromosome 14 open reading frame 144 |
| chr11_-_132813566 | 0.13 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr1_+_209859510 | 0.13 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr4_-_85771168 | 0.12 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr22_+_35462129 | 0.12 |
ENST00000404699.2
ENST00000308700.6 |
ISX
|
intestine-specific homeobox |
| chr2_+_101591314 | 0.12 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr12_+_93096619 | 0.12 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr4_-_299110 | 0.12 |
ENST00000419098.1
|
ZNF732
|
zinc finger protein 732 |
| chr2_-_216878305 | 0.12 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr9_+_130911770 | 0.12 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr16_+_2285817 | 0.11 |
ENST00000564065.1
|
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr18_+_61554932 | 0.11 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chrX_-_133792480 | 0.11 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr21_+_39644214 | 0.11 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_-_100643765 | 0.11 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr7_+_141418962 | 0.11 |
ENST00000493845.1
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr5_+_142286887 | 0.11 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr13_+_78315528 | 0.11 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_-_12679171 | 0.11 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr12_+_59989918 | 0.11 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr8_+_119294456 | 0.11 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr1_+_229440129 | 0.11 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr10_+_69865866 | 0.11 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr8_+_35649365 | 0.11 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chrM_+_14741 | 0.11 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr21_+_39644305 | 0.11 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr17_-_39165366 | 0.11 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr11_-_47447970 | 0.11 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr14_+_105864847 | 0.10 |
ENST00000451127.2
|
TEX22
|
testis expressed 22 |
| chrM_+_12331 | 0.10 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr15_-_71184724 | 0.10 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr2_+_135596180 | 0.10 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr20_+_52105495 | 0.10 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr6_+_26156551 | 0.10 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr4_+_117220016 | 0.10 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr21_+_39644395 | 0.10 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr7_-_27205136 | 0.10 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr11_-_102714534 | 0.10 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr9_-_133814455 | 0.10 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr22_-_38240316 | 0.10 |
ENST00000411961.2
ENST00000434930.1 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr17_-_38928414 | 0.10 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr2_+_200472779 | 0.10 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr20_-_52612468 | 0.10 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr1_+_6640046 | 0.10 |
ENST00000319084.5
ENST00000435905.1 |
ZBTB48
|
zinc finger and BTB domain containing 48 |
| chr22_-_30814469 | 0.10 |
ENST00000598426.1
|
KIAA1658
|
KIAA1658 |
| chr4_+_124571409 | 0.10 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr1_+_66458072 | 0.10 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr8_-_95220775 | 0.10 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr17_-_47786375 | 0.10 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr9_-_130889990 | 0.10 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr17_+_22022437 | 0.10 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr20_+_62327996 | 0.10 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr15_+_67841330 | 0.09 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr11_-_26744908 | 0.09 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr21_+_39644172 | 0.09 |
ENST00000398932.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr12_+_93096759 | 0.09 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr17_+_7533439 | 0.09 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr2_-_165424973 | 0.09 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr9_+_105757590 | 0.09 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr18_+_29769978 | 0.09 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr12_-_56236734 | 0.09 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr6_-_133079022 | 0.09 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr9_+_44868935 | 0.09 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr6_-_116833500 | 0.09 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr13_+_78315466 | 0.09 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr10_+_48189612 | 0.08 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr17_+_61473104 | 0.08 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr3_+_160394940 | 0.08 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr14_+_21492331 | 0.08 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr5_-_111093759 | 0.08 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr3_+_15045419 | 0.08 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr7_-_50860565 | 0.08 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
| chrX_-_55020511 | 0.08 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_-_148939598 | 0.08 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr4_-_68749699 | 0.08 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr1_-_152386732 | 0.08 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr20_-_52687030 | 0.08 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr4_+_39046615 | 0.08 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr2_-_70417827 | 0.08 |
ENST00000457952.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr6_-_27114577 | 0.08 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr6_+_12007897 | 0.08 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr5_-_88119580 | 0.08 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr15_-_41166414 | 0.08 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr5_+_61874562 | 0.08 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr10_+_75545391 | 0.07 |
ENST00000604524.1
ENST00000605216.1 ENST00000398706.2 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr15_+_67418047 | 0.07 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr19_+_42041860 | 0.07 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr19_+_49468558 | 0.07 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr5_-_43412418 | 0.07 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr3_-_112127981 | 0.07 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr10_+_75545329 | 0.07 |
ENST00000604729.1
ENST00000603114.1 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr11_-_76155618 | 0.07 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr11_+_5775923 | 0.07 |
ENST00000317254.3
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4 (gene/pseudogene) |
| chr16_-_24942273 | 0.07 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr3_+_30648066 | 0.07 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr3_+_151531859 | 0.07 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr15_-_78538025 | 0.07 |
ENST00000560817.1
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr17_-_39140549 | 0.07 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr6_+_30687978 | 0.07 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr14_-_69444957 | 0.07 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr1_-_43919573 | 0.07 |
ENST00000372432.1
ENST00000372425.4 ENST00000583037.1 |
HYI
|
hydroxypyruvate isomerase (putative) |
| chr8_+_77593448 | 0.07 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr5_-_13944652 | 0.07 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr11_+_117070037 | 0.07 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr15_+_45938079 | 0.07 |
ENST00000561493.1
|
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr3_-_15563229 | 0.07 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr3_-_141747950 | 0.07 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_57326704 | 0.06 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr6_+_131958436 | 0.06 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chrM_-_14670 | 0.06 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr2_+_90248739 | 0.06 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr22_-_38240412 | 0.06 |
ENST00000215941.4
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr2_+_176994408 | 0.06 |
ENST00000429017.1
ENST00000313173.4 ENST00000544999.1 |
HOXD8
|
homeobox D8 |
| chr10_+_35456444 | 0.06 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr7_+_16793160 | 0.06 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr3_+_185046676 | 0.06 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr19_+_18303992 | 0.06 |
ENST00000599612.2
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr17_+_46918925 | 0.06 |
ENST00000502761.1
|
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr20_+_3776371 | 0.06 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr1_-_209957882 | 0.06 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr12_-_52761262 | 0.06 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr3_-_114790179 | 0.06 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr17_-_41132410 | 0.06 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr17_+_61086917 | 0.06 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr19_-_54619006 | 0.06 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr10_+_133747955 | 0.06 |
ENST00000455566.1
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr11_-_2158507 | 0.06 |
ENST00000381392.1
ENST00000381395.1 ENST00000418738.2 |
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr15_+_64680003 | 0.06 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr9_-_133814527 | 0.06 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr2_+_27440229 | 0.06 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr11_-_47447767 | 0.06 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr4_-_184241927 | 0.06 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr17_-_41739283 | 0.06 |
ENST00000393661.2
ENST00000318579.4 |
MEOX1
|
mesenchyme homeobox 1 |
| chr1_-_86848760 | 0.06 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr7_-_27219632 | 0.06 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr14_-_89878369 | 0.05 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr12_-_54071181 | 0.05 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr17_-_2117600 | 0.05 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr6_+_88054530 | 0.05 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr16_+_58035277 | 0.05 |
ENST00000219281.3
ENST00000539737.2 ENST00000423271.3 ENST00000561568.1 ENST00000563149.1 |
USB1
|
U6 snRNA biogenesis 1 |
| chr3_+_70048881 | 0.05 |
ENST00000483525.1
|
RP11-460N16.1
|
RP11-460N16.1 |
| chr16_+_30662085 | 0.05 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr13_-_44735393 | 0.05 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr1_+_151735431 | 0.05 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr8_-_60031762 | 0.05 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr13_+_23755054 | 0.05 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC10_HOXD13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 0.6 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.9 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) |
| 0.1 | 0.4 | GO:0060584 | nitric oxide transport(GO:0030185) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.2 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:1904478 | regulation of intestinal absorption(GO:1904478) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.0 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.0 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.0 | 0.0 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 0.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 1.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.0 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.0 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.0 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |