Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
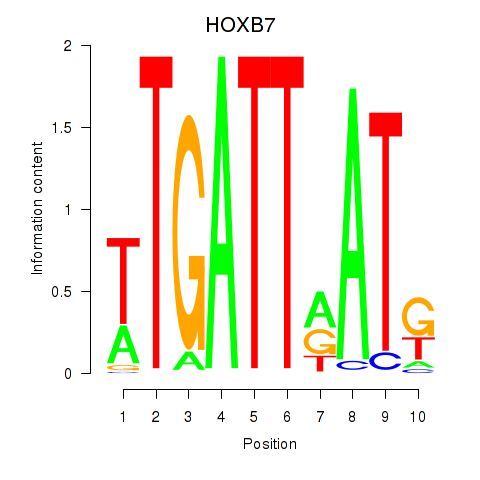
Results for HOXB7
Z-value: 0.74
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | 0.33 | 5.2e-01 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_131409476 | 0.64 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr10_-_106240032 | 0.48 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr7_-_33842742 | 0.37 |
ENST00000420185.1
ENST00000440034.1 |
RP11-89N17.4
|
RP11-89N17.4 |
| chr12_-_86650077 | 0.34 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr12_+_21679220 | 0.33 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr11_-_57194817 | 0.32 |
ENST00000529748.1
ENST00000525474.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr19_+_12203100 | 0.32 |
ENST00000596883.1
|
ZNF788
|
zinc finger family member 788 |
| chr11_+_18154059 | 0.31 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr4_+_71384257 | 0.31 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr4_+_71384300 | 0.30 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr19_+_35417939 | 0.29 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr9_+_74729511 | 0.28 |
ENST00000545168.1
|
GDA
|
guanine deaminase |
| chr10_+_69865866 | 0.28 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr17_+_70036164 | 0.28 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr6_+_130339710 | 0.28 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr4_-_85771168 | 0.26 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr4_+_26324474 | 0.26 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr22_-_30642728 | 0.25 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr12_-_91546926 | 0.25 |
ENST00000550758.1
|
DCN
|
decorin |
| chr22_-_30642782 | 0.24 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr16_+_77756399 | 0.24 |
ENST00000564085.1
ENST00000268533.5 ENST00000568787.1 ENST00000437314.3 ENST00000563839.1 |
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr19_+_12203069 | 0.23 |
ENST00000430298.2
ENST00000339302.4 |
ZNF788
ZNF788
|
zinc finger family member 788 Zinc finger protein 788 |
| chr8_-_91618285 | 0.23 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr3_+_177545563 | 0.23 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr10_-_51958906 | 0.21 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr19_+_35417844 | 0.21 |
ENST00000601957.1
|
ZNF30
|
zinc finger protein 30 |
| chr4_-_186570679 | 0.21 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_48774586 | 0.20 |
ENST00000594024.1
ENST00000595408.1 ENST00000315849.1 |
ZNF114
|
zinc finger protein 114 |
| chr4_+_158493642 | 0.20 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr1_-_92952433 | 0.20 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr15_-_28957469 | 0.20 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr11_-_2162162 | 0.19 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr11_+_44117219 | 0.19 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr2_-_208030295 | 0.19 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chrX_+_1387693 | 0.18 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr12_-_7596735 | 0.18 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr15_+_64680003 | 0.18 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr18_+_61554932 | 0.17 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr17_-_69198295 | 0.17 |
ENST00000569074.1
|
CASC17
|
cancer susceptibility candidate 17 (non-protein coding) |
| chr16_-_66583994 | 0.17 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr3_-_141747950 | 0.17 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_224624730 | 0.17 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr19_+_35417798 | 0.17 |
ENST00000303586.7
ENST00000439785.1 ENST00000601540.1 |
ZNF30
|
zinc finger protein 30 |
| chr1_-_206288647 | 0.16 |
ENST00000331555.5
|
C1orf186
|
chromosome 1 open reading frame 186 |
| chr12_-_95510743 | 0.16 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr16_-_66583701 | 0.16 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr19_+_37837218 | 0.16 |
ENST00000591134.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr2_+_166095898 | 0.16 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr3_+_173116225 | 0.16 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr5_+_140227357 | 0.16 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr2_-_70780770 | 0.16 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr21_-_31588365 | 0.15 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr20_+_25388293 | 0.15 |
ENST00000262460.4
ENST00000429262.2 |
GINS1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr14_-_34931458 | 0.15 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr18_+_22040593 | 0.15 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr12_+_96252706 | 0.15 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr16_-_30122717 | 0.15 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr11_+_44117099 | 0.15 |
ENST00000533608.1
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr11_+_44117260 | 0.15 |
ENST00000358681.4
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr17_-_4938712 | 0.15 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr18_+_20714525 | 0.14 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr13_-_113242439 | 0.14 |
ENST00000375669.3
ENST00000261965.3 |
TUBGCP3
|
tubulin, gamma complex associated protein 3 |
| chr22_-_22337154 | 0.14 |
ENST00000413067.2
ENST00000437929.1 ENST00000456075.1 ENST00000434517.1 ENST00000424393.1 ENST00000449704.1 ENST00000437103.1 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr2_-_203735484 | 0.14 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr2_+_120436760 | 0.14 |
ENST00000445518.1
ENST00000409951.1 |
TMEM177
|
transmembrane protein 177 |
| chr10_+_90484301 | 0.13 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr16_-_66584059 | 0.13 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr6_+_89674246 | 0.13 |
ENST00000369474.1
|
AL079342.1
|
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr4_+_36283213 | 0.13 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr7_-_120498357 | 0.13 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr16_-_65106110 | 0.13 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr14_+_55034599 | 0.13 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr13_-_103719196 | 0.13 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr7_+_117824086 | 0.13 |
ENST00000249299.2
ENST00000424702.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr12_+_52056548 | 0.13 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr1_-_91487806 | 0.13 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr12_-_118628315 | 0.13 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr15_-_32695396 | 0.13 |
ENST00000512626.2
ENST00000435655.2 |
GOLGA8K
AC139426.2
|
golgin A8 family, member K Uncharacterized protein; cDNA FLJ52611 |
| chr15_-_23692381 | 0.13 |
ENST00000567107.1
ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2
|
golgin A6 family-like 2 |
| chr16_+_15596123 | 0.13 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr12_+_123944070 | 0.13 |
ENST00000412157.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr8_-_37411648 | 0.13 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr8_+_31497271 | 0.12 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr9_-_70465758 | 0.12 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr11_-_8795787 | 0.12 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr20_-_1309809 | 0.12 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr20_-_50179368 | 0.12 |
ENST00000609943.1
ENST00000609507.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr18_-_73967160 | 0.12 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr3_+_101659682 | 0.12 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr16_-_67427389 | 0.12 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr15_-_60683326 | 0.12 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr2_+_27886330 | 0.11 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr12_-_39734783 | 0.11 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr7_-_7782204 | 0.11 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr3_+_157827841 | 0.11 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr11_+_57365150 | 0.11 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr8_-_29208183 | 0.11 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr10_+_102891048 | 0.11 |
ENST00000467928.2
|
TLX1
|
T-cell leukemia homeobox 1 |
| chr16_-_66584015 | 0.11 |
ENST00000545043.2
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr21_-_33984456 | 0.11 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr19_+_52693259 | 0.11 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr3_+_136581096 | 0.11 |
ENST00000476286.1
ENST00000488930.1 |
NCK1
|
NCK adaptor protein 1 |
| chr11_-_102668879 | 0.11 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr3_+_158787041 | 0.11 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr15_+_86098670 | 0.11 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr2_+_234600253 | 0.11 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr17_+_22022437 | 0.11 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr3_+_159557637 | 0.11 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr15_+_75575176 | 0.11 |
ENST00000434739.3
|
GOLGA6D
|
golgin A6 family, member D |
| chr13_-_46756351 | 0.10 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr14_-_101295407 | 0.10 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr8_-_109799793 | 0.10 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr12_+_8309630 | 0.10 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr12_+_59989791 | 0.10 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr14_+_74417192 | 0.10 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr1_-_165414414 | 0.10 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr9_+_140135665 | 0.10 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr3_+_140947563 | 0.10 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr15_+_28623784 | 0.10 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr3_+_139063372 | 0.10 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr12_-_47226152 | 0.10 |
ENST00000546940.1
|
SLC38A4
|
solute carrier family 38, member 4 |
| chr10_+_118083919 | 0.10 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr1_+_152956549 | 0.10 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr7_-_86595190 | 0.10 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr15_-_28778117 | 0.10 |
ENST00000525590.2
ENST00000329523.6 |
GOLGA8G
|
golgin A8 family, member G |
| chr8_-_116681123 | 0.10 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr10_+_69869237 | 0.10 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr5_-_10761206 | 0.10 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr1_-_209957882 | 0.10 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr17_+_45973516 | 0.10 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr19_-_48753064 | 0.10 |
ENST00000520153.1
ENST00000357778.5 ENST00000520015.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr3_+_52570610 | 0.10 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr9_-_85882145 | 0.10 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr14_+_103851712 | 0.09 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr12_-_28124903 | 0.09 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr12_-_11139511 | 0.09 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr3_-_4927447 | 0.09 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr3_-_24207039 | 0.09 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr2_+_44396000 | 0.09 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr1_+_66820058 | 0.09 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr3_+_156009623 | 0.09 |
ENST00000389634.5
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr6_-_116866773 | 0.09 |
ENST00000368602.3
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr17_+_7477040 | 0.09 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr8_-_10512569 | 0.09 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr19_-_20748614 | 0.09 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chr8_-_116681686 | 0.09 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr11_+_71927807 | 0.09 |
ENST00000298223.6
ENST00000454954.2 ENST00000541003.1 ENST00000539412.1 ENST00000536778.1 ENST00000535625.1 ENST00000321324.7 |
FOLR2
|
folate receptor 2 (fetal) |
| chr7_-_138363824 | 0.09 |
ENST00000419765.3
|
SVOPL
|
SVOP-like |
| chr16_+_20911174 | 0.09 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr9_+_131267052 | 0.09 |
ENST00000539582.1
|
GLE1
|
GLE1 RNA export mediator |
| chr7_-_105926058 | 0.09 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr15_+_30375158 | 0.09 |
ENST00000341650.6
ENST00000567927.1 |
GOLGA8J
|
golgin A8 family, member J |
| chr4_+_71600144 | 0.09 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr15_+_67418047 | 0.09 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr2_-_230787879 | 0.09 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr17_+_41363854 | 0.08 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr6_-_15548591 | 0.08 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr9_+_77112244 | 0.08 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr2_+_44001172 | 0.08 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr1_-_20503917 | 0.08 |
ENST00000429261.2
|
PLA2G2C
|
phospholipase A2, group IIC |
| chr3_-_139396787 | 0.08 |
ENST00000296202.7
ENST00000509291.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr16_+_82660635 | 0.08 |
ENST00000567445.1
ENST00000446376.2 |
CDH13
|
cadherin 13 |
| chrX_-_110655391 | 0.08 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr11_-_107729287 | 0.08 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_-_69434245 | 0.08 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr5_+_125759140 | 0.08 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr1_+_244515930 | 0.08 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr8_-_67874805 | 0.08 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr10_+_124320156 | 0.08 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr14_+_24584056 | 0.08 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr8_+_68864330 | 0.08 |
ENST00000288368.4
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr7_+_116654935 | 0.08 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr19_+_44645700 | 0.08 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr16_+_67880574 | 0.08 |
ENST00000219169.4
|
NUTF2
|
nuclear transport factor 2 |
| chr17_-_38821373 | 0.08 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr14_+_24584372 | 0.08 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr1_-_9953295 | 0.08 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr1_-_40562908 | 0.08 |
ENST00000527311.2
ENST00000449045.2 ENST00000372779.4 |
PPT1
|
palmitoyl-protein thioesterase 1 |
| chr15_-_32747835 | 0.08 |
ENST00000509311.2
ENST00000414865.2 |
GOLGA8O
|
golgin A8 family, member O |
| chr15_+_43477455 | 0.08 |
ENST00000300213.4
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chrX_+_1734051 | 0.08 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr19_-_49955050 | 0.08 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr4_-_186734275 | 0.08 |
ENST00000456060.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_+_56401268 | 0.08 |
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr6_-_74231444 | 0.08 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chrM_+_9207 | 0.07 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr2_-_74735707 | 0.07 |
ENST00000233630.6
|
PCGF1
|
polycomb group ring finger 1 |
| chr6_-_90025011 | 0.07 |
ENST00000402938.3
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr4_-_177116772 | 0.07 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr7_-_107883678 | 0.07 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr19_+_41698927 | 0.07 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr2_-_43266680 | 0.07 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr19_+_38880252 | 0.07 |
ENST00000586301.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chrX_-_110655306 | 0.07 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr10_-_128975273 | 0.07 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr12_-_109025849 | 0.07 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr14_+_35747825 | 0.07 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr12_+_54402790 | 0.07 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr12_+_9980069 | 0.07 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr12_-_54653313 | 0.07 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.6 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.3 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.6 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.2 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.2 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.3 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0052416 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:0071231 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0048858 | cell projection morphogenesis(GO:0048858) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0046549 | amacrine cell differentiation(GO:0035881) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.0 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.3 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.0 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.6 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.5 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.3 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |