Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
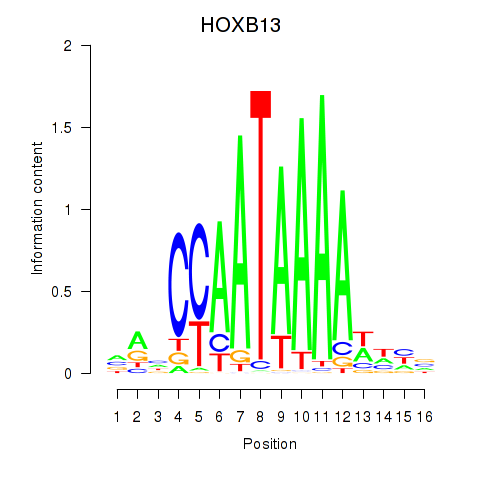
Results for HOXB13
Z-value: 0.37
Transcription factors associated with HOXB13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB13
|
ENSG00000159184.7 | homeobox B13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB13 | hg19_v2_chr17_-_46806540_46806558 | 0.93 | 7.4e-03 | Click! |
Activity profile of HOXB13 motif
Sorted Z-values of HOXB13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_27782548 | 0.22 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr1_-_12679171 | 0.21 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr16_-_2205352 | 0.19 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr17_-_46806540 | 0.16 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chrM_-_14670 | 0.13 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr18_-_73967160 | 0.13 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr17_-_10560619 | 0.11 |
ENST00000583535.1
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr22_-_38240316 | 0.10 |
ENST00000411961.2
ENST00000434930.1 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr17_-_73663168 | 0.10 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr3_-_195310802 | 0.10 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr4_+_110749143 | 0.10 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chrM_+_12331 | 0.10 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrM_+_14741 | 0.09 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr7_+_128784712 | 0.09 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr14_-_50583271 | 0.09 |
ENST00000395860.2
ENST00000395859.2 |
VCPKMT
|
valosin containing protein lysine (K) methyltransferase |
| chr5_-_126409159 | 0.09 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr22_-_38240412 | 0.08 |
ENST00000215941.4
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr9_+_44868935 | 0.07 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr8_+_97597148 | 0.07 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr3_-_114790179 | 0.07 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_+_30669720 | 0.07 |
ENST00000356166.6
|
FBRS
|
fibrosin |
| chr16_-_71610985 | 0.07 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr14_-_68000442 | 0.07 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr17_-_73663245 | 0.06 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr3_+_89156799 | 0.06 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr17_+_18086392 | 0.06 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr11_-_102714534 | 0.06 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr1_-_45956868 | 0.05 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr11_+_118401706 | 0.05 |
ENST00000411589.2
ENST00000442938.2 ENST00000359862.4 |
TMEM25
|
transmembrane protein 25 |
| chr1_-_54879140 | 0.04 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr9_-_107367951 | 0.04 |
ENST00000542196.1
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2 |
| chr14_+_76776957 | 0.04 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr9_+_22646189 | 0.04 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr6_-_116833500 | 0.04 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr19_+_35634146 | 0.04 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr18_-_52989217 | 0.03 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr1_-_45956800 | 0.03 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr4_+_169575875 | 0.03 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr6_-_43021612 | 0.03 |
ENST00000535468.1
|
CUL7
|
cullin 7 |
| chr11_-_102826434 | 0.03 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr22_+_35462129 | 0.03 |
ENST00000404699.2
ENST00000308700.6 |
ISX
|
intestine-specific homeobox |
| chr4_-_69111401 | 0.03 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr7_-_11871815 | 0.03 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr1_+_23037323 | 0.02 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr1_+_78769549 | 0.02 |
ENST00000370758.1
|
PTGFR
|
prostaglandin F receptor (FP) |
| chr8_-_114449112 | 0.02 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr15_+_71839566 | 0.02 |
ENST00000357769.4
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr1_-_45956822 | 0.02 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr13_+_28813645 | 0.02 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr18_+_68002675 | 0.02 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr21_+_30673091 | 0.02 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr9_+_125132803 | 0.02 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr6_-_139613269 | 0.02 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr16_+_2205755 | 0.02 |
ENST00000326181.6
|
TRAF7
|
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
| chr9_-_14180778 | 0.02 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr8_+_77593448 | 0.02 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr1_-_149783914 | 0.02 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr9_+_125795788 | 0.01 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr12_+_8309630 | 0.01 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr2_-_175629135 | 0.01 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_+_66458072 | 0.01 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr15_-_55541227 | 0.01 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_+_89156674 | 0.01 |
ENST00000336596.2
|
EPHA3
|
EPH receptor A3 |
| chr1_-_36863481 | 0.01 |
ENST00000315732.2
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr9_+_123884038 | 0.01 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr9_+_125133315 | 0.01 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr2_-_136288113 | 0.01 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr16_-_24942273 | 0.01 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr12_+_70219052 | 0.01 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr2_+_74685527 | 0.01 |
ENST00000393972.3
ENST00000409737.1 ENST00000428943.1 |
WBP1
|
WW domain binding protein 1 |
| chr3_-_123710199 | 0.01 |
ENST00000184183.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr15_+_66994561 | 0.01 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr16_-_24942411 | 0.01 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr8_+_35649365 | 0.01 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr1_-_155880672 | 0.01 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr8_+_119294456 | 0.00 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr1_+_52682052 | 0.00 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr13_-_45768841 | 0.00 |
ENST00000379108.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr9_-_104198042 | 0.00 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr9_+_103947311 | 0.00 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr13_-_103719196 | 0.00 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr2_-_51259292 | 0.00 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr8_+_77593474 | 0.00 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr7_-_50860565 | 0.00 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.0 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |