Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
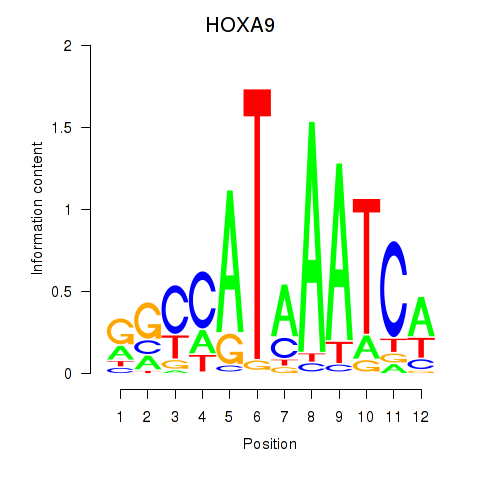
Results for HOXA9
Z-value: 0.52
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.11 | homeobox A9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg19_v2_chr7_-_27205136_27205164 | -0.19 | 7.2e-01 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_25281747 | 0.36 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr18_+_61564389 | 0.35 |
ENST00000397996.2
ENST00000418725.1 |
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr8_-_65730127 | 0.32 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr1_-_204135450 | 0.29 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr4_-_74964904 | 0.26 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr1_+_209545365 | 0.25 |
ENST00000447257.1
|
RP11-372M18.2
|
RP11-372M18.2 |
| chr2_-_214017151 | 0.23 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_+_149230680 | 0.22 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chr6_-_27775694 | 0.21 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr8_-_37411648 | 0.20 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr12_-_59175485 | 0.20 |
ENST00000550678.1
ENST00000552201.1 |
RP11-767I20.1
|
RP11-767I20.1 |
| chr7_-_75241096 | 0.20 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr2_-_114461655 | 0.19 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr18_-_73967160 | 0.19 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr2_+_138721850 | 0.19 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr6_+_143447322 | 0.18 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr7_-_83824449 | 0.18 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_-_224467002 | 0.17 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr5_+_147582348 | 0.16 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr16_+_70613770 | 0.16 |
ENST00000429149.2
ENST00000563721.2 |
IL34
|
interleukin 34 |
| chr1_+_38512799 | 0.16 |
ENST00000432922.1
ENST00000428151.1 |
RP5-884C9.2
|
RP5-884C9.2 |
| chr4_+_169633310 | 0.16 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr16_-_67427389 | 0.15 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr16_-_55867146 | 0.15 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr17_+_61473104 | 0.15 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_-_152552980 | 0.15 |
ENST00000368787.3
|
LCE3D
|
late cornified envelope 3D |
| chr10_+_67330096 | 0.15 |
ENST00000433152.4
ENST00000601979.1 ENST00000599409.1 ENST00000608678.1 |
RP11-222A11.1
|
RP11-222A11.1 |
| chr21_-_31852663 | 0.14 |
ENST00000390689.2
|
KRTAP19-1
|
keratin associated protein 19-1 |
| chr22_-_38240316 | 0.14 |
ENST00000411961.2
ENST00000434930.1 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr5_-_94417339 | 0.13 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr3_+_142842128 | 0.13 |
ENST00000483262.1
|
RP11-80H8.4
|
RP11-80H8.4 |
| chr3_-_52864680 | 0.13 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr17_+_4853442 | 0.13 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr16_+_15596123 | 0.13 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr20_+_32250079 | 0.13 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chr3_+_177545563 | 0.12 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr2_+_219646462 | 0.12 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr3_-_108672742 | 0.12 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr3_+_157827841 | 0.12 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr8_+_118532937 | 0.11 |
ENST00000297347.3
|
MED30
|
mediator complex subunit 30 |
| chr7_-_7679633 | 0.11 |
ENST00000401447.1
|
RPA3
|
replication protein A3, 14kDa |
| chr18_+_9102669 | 0.11 |
ENST00000497577.2
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr18_-_67624412 | 0.11 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr16_+_56691606 | 0.11 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr5_-_137374288 | 0.11 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr6_-_110011704 | 0.11 |
ENST00000448084.2
|
AK9
|
adenylate kinase 9 |
| chr5_-_168006324 | 0.11 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr5_+_176731572 | 0.11 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr6_-_133035185 | 0.11 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr5_+_147582387 | 0.10 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr3_-_108672609 | 0.10 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr10_-_96829246 | 0.10 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr17_+_73663470 | 0.10 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
| chrX_-_63615297 | 0.10 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr22_+_23248512 | 0.10 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr2_-_224467093 | 0.09 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr5_+_177433406 | 0.09 |
ENST00000504530.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr17_-_48450534 | 0.09 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr2_-_31637560 | 0.09 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr12_+_122181529 | 0.09 |
ENST00000541467.1
|
TMEM120B
|
transmembrane protein 120B |
| chr10_-_101190202 | 0.09 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr4_-_76944621 | 0.09 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr19_+_45971246 | 0.09 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr22_-_30642782 | 0.09 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr5_+_38845960 | 0.09 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr17_-_29151686 | 0.09 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr1_+_44584522 | 0.09 |
ENST00000372299.3
|
KLF17
|
Kruppel-like factor 17 |
| chr14_-_81425828 | 0.09 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr12_-_371994 | 0.09 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr10_-_51958906 | 0.08 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr16_+_19079215 | 0.08 |
ENST00000544894.2
ENST00000561858.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr21_-_30257669 | 0.08 |
ENST00000303775.5
ENST00000351429.3 |
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr14_+_88490894 | 0.08 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr2_-_99870744 | 0.08 |
ENST00000409238.1
ENST00000423800.1 |
LYG2
|
lysozyme G-like 2 |
| chr1_-_23340400 | 0.08 |
ENST00000440767.2
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr19_+_4769117 | 0.08 |
ENST00000540211.1
ENST00000317292.3 ENST00000586721.1 ENST00000592709.1 ENST00000588711.1 ENST00000589639.1 ENST00000591008.1 ENST00000592663.1 ENST00000588758.1 |
MIR7-3HG
|
MIR7-3 host gene (non-protein coding) |
| chr8_+_31497271 | 0.08 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr11_+_113185778 | 0.08 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr1_-_43282945 | 0.08 |
ENST00000537227.1
|
CCDC23
|
coiled-coil domain containing 23 |
| chr16_+_67063855 | 0.08 |
ENST00000563939.2
|
CBFB
|
core-binding factor, beta subunit |
| chr9_+_44867571 | 0.08 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr12_+_122880045 | 0.08 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chrX_+_70503526 | 0.08 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr1_-_153085984 | 0.08 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chr11_+_5646213 | 0.08 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr2_+_32502952 | 0.08 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr8_+_42128861 | 0.08 |
ENST00000518983.1
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr11_+_59522837 | 0.08 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr16_-_18462221 | 0.07 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr12_+_59989918 | 0.07 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_+_108994466 | 0.07 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr6_-_110011718 | 0.07 |
ENST00000532976.1
|
AK9
|
adenylate kinase 9 |
| chr12_+_57853918 | 0.07 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr7_-_27169801 | 0.07 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr16_-_90128733 | 0.07 |
ENST00000325921.6
|
PRDM7
|
PR domain containing 7 |
| chr8_-_91618285 | 0.07 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr11_-_3400442 | 0.07 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr7_+_16793160 | 0.07 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr18_+_3466248 | 0.07 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr4_-_186732241 | 0.07 |
ENST00000421639.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_73996987 | 0.07 |
ENST00000588812.1
ENST00000448471.1 |
CDK3
|
cyclin-dependent kinase 3 |
| chr3_+_158787041 | 0.07 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr20_-_36661826 | 0.07 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr12_+_24857397 | 0.07 |
ENST00000536131.1
|
RP11-625L16.1
|
RP11-625L16.1 |
| chr4_+_71384300 | 0.06 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr7_-_99332719 | 0.06 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr19_-_47975417 | 0.06 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr17_+_73663402 | 0.06 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr3_+_57875711 | 0.06 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr3_-_100558953 | 0.06 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr10_-_71176623 | 0.06 |
ENST00000373306.4
|
TACR2
|
tachykinin receptor 2 |
| chr1_-_205819245 | 0.06 |
ENST00000367136.4
|
PM20D1
|
peptidase M20 domain containing 1 |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr7_-_33842742 | 0.06 |
ENST00000420185.1
ENST00000440034.1 |
RP11-89N17.4
|
RP11-89N17.4 |
| chr17_-_7232585 | 0.06 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr2_+_44396000 | 0.06 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr18_+_11857439 | 0.06 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr5_+_80529104 | 0.06 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr6_-_55443831 | 0.06 |
ENST00000428842.1
ENST00000358072.5 ENST00000508459.1 |
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr11_+_71927807 | 0.06 |
ENST00000298223.6
ENST00000454954.2 ENST00000541003.1 ENST00000539412.1 ENST00000536778.1 ENST00000535625.1 ENST00000321324.7 |
FOLR2
|
folate receptor 2 (fetal) |
| chr3_-_151176497 | 0.06 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr6_+_90272339 | 0.06 |
ENST00000522779.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr2_+_238600933 | 0.06 |
ENST00000420665.1
ENST00000392000.4 |
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr22_-_31324215 | 0.06 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr8_+_24151553 | 0.06 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr9_+_77112244 | 0.06 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr6_-_131211534 | 0.05 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr15_-_41166414 | 0.05 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr6_+_90272488 | 0.05 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr7_+_153584166 | 0.05 |
ENST00000404039.1
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr17_-_8021710 | 0.05 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr11_-_59952106 | 0.05 |
ENST00000529054.1
ENST00000530839.1 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr19_-_50464338 | 0.05 |
ENST00000426971.2
ENST00000447370.2 |
SIGLEC11
|
sialic acid binding Ig-like lectin 11 |
| chr2_+_13677795 | 0.05 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr11_+_101918153 | 0.05 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr16_+_3074002 | 0.05 |
ENST00000326266.8
ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6
|
THO complex 6 homolog (Drosophila) |
| chr1_+_150254936 | 0.05 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr15_+_59730348 | 0.05 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr1_-_144866711 | 0.05 |
ENST00000530130.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr10_+_96698406 | 0.05 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr17_+_39421591 | 0.05 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr16_-_65106110 | 0.05 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr14_-_24911448 | 0.05 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr15_+_67418047 | 0.05 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr18_-_70532906 | 0.05 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr10_+_91092241 | 0.05 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr5_-_94417314 | 0.05 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr3_-_48130707 | 0.05 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr7_+_104581389 | 0.05 |
ENST00000415513.1
ENST00000417026.1 |
RP11-325F22.2
|
RP11-325F22.2 |
| chr19_+_44645700 | 0.05 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr5_+_38846101 | 0.05 |
ENST00000274276.3
|
OSMR
|
oncostatin M receptor |
| chr1_-_234614849 | 0.05 |
ENST00000040877.1
|
TARBP1
|
TAR (HIV-1) RNA binding protein 1 |
| chr19_+_50529212 | 0.05 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chr11_-_76155618 | 0.05 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr16_+_70557685 | 0.05 |
ENST00000302516.5
ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3
|
splicing factor 3b, subunit 3, 130kDa |
| chr12_-_3982548 | 0.05 |
ENST00000397096.2
ENST00000447133.3 ENST00000450737.2 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr1_-_92951607 | 0.05 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr16_+_16434185 | 0.05 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chr10_+_5488564 | 0.05 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr1_-_24513737 | 0.05 |
ENST00000374421.3
ENST00000374418.3 ENST00000327535.1 ENST00000327575.2 |
IFNLR1
|
interferon, lambda receptor 1 |
| chr14_+_92789498 | 0.05 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr11_-_2162162 | 0.05 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr14_-_74960030 | 0.05 |
ENST00000553490.1
ENST00000557510.1 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr17_+_54230819 | 0.05 |
ENST00000318698.2
ENST00000566473.2 |
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr5_-_180288248 | 0.05 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr8_-_10512569 | 0.04 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr5_-_175388327 | 0.04 |
ENST00000432305.2
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr10_+_24755416 | 0.04 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr11_+_107650219 | 0.04 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr2_-_130031335 | 0.04 |
ENST00000375987.3
|
AC079586.1
|
AC079586.1 |
| chr15_+_45315302 | 0.04 |
ENST00000267814.9
|
SORD
|
sorbitol dehydrogenase |
| chr1_-_207095324 | 0.04 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr1_-_33502441 | 0.04 |
ENST00000548033.1
ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2
|
adenylate kinase 2 |
| chr12_-_122879969 | 0.04 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr8_-_28347737 | 0.04 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr8_-_67874805 | 0.04 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr10_+_102222798 | 0.04 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr6_-_55443958 | 0.04 |
ENST00000370850.2
|
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr1_-_43919346 | 0.04 |
ENST00000372430.3
ENST00000372433.1 ENST00000372434.1 ENST00000486909.1 |
HYI
|
hydroxypyruvate isomerase (putative) |
| chr1_-_79472365 | 0.04 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr8_-_116673894 | 0.04 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_+_47666275 | 0.04 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr6_-_74231444 | 0.04 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr5_-_131347306 | 0.04 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr8_+_38261880 | 0.04 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr5_-_94417186 | 0.04 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chrX_-_130037162 | 0.04 |
ENST00000432489.1
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chrX_-_130037198 | 0.04 |
ENST00000370935.1
ENST00000338144.3 ENST00000394363.1 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr6_-_131299929 | 0.04 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr12_+_25205628 | 0.04 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chrX_+_70503433 | 0.04 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr15_+_59664884 | 0.04 |
ENST00000558348.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr16_+_72088376 | 0.04 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr8_+_42128812 | 0.04 |
ENST00000520810.1
ENST00000416505.2 ENST00000519735.1 ENST00000520835.1 ENST00000379708.3 |
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr2_+_113033164 | 0.04 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr13_+_98794810 | 0.04 |
ENST00000595437.1
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr14_+_78174414 | 0.04 |
ENST00000557342.1
ENST00000238688.5 ENST00000557623.1 ENST00000557431.1 ENST00000556831.1 ENST00000556375.1 ENST00000553981.1 |
SLIRP
|
SRA stem-loop interacting RNA binding protein |
| chr11_-_35441597 | 0.04 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_+_47293795 | 0.04 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr19_+_50528971 | 0.04 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.2 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.2 | GO:0061569 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.3 | GO:0042756 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) drinking behavior(GO:0042756) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0071231 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.0 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.0 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |