Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
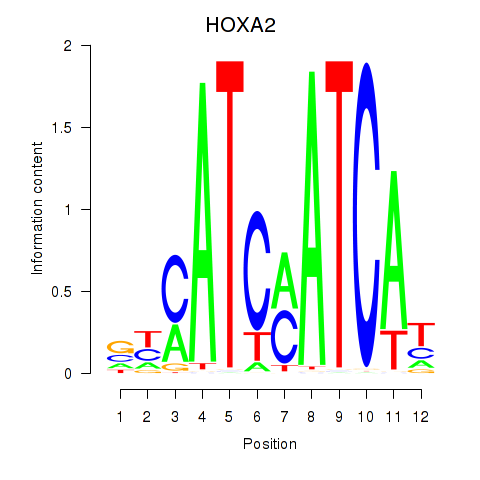
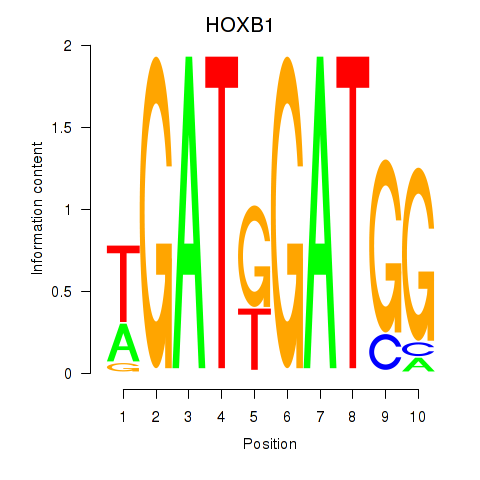
Results for HOXA2_HOXB1
Z-value: 0.63
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA2
|
ENSG00000105996.5 | homeobox A2 |
|
HOXB1
|
ENSG00000120094.6 | homeobox B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | -0.92 | 9.0e-03 | Click! |
| HOXB1 | hg19_v2_chr17_-_46608272_46608385 | 0.35 | 4.9e-01 | Click! |
Activity profile of HOXA2_HOXB1 motif
Sorted Z-values of HOXA2_HOXB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_7758374 | 0.40 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr20_+_52105495 | 0.35 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr11_-_126174186 | 0.32 |
ENST00000524964.1
|
RP11-712L6.5
|
Uncharacterized protein |
| chr12_-_15038779 | 0.24 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr5_+_147582348 | 0.22 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr16_-_67427389 | 0.22 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr14_-_21516590 | 0.19 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr10_+_102891048 | 0.19 |
ENST00000467928.2
|
TLX1
|
T-cell leukemia homeobox 1 |
| chr7_-_143956815 | 0.19 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr17_-_57229155 | 0.19 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr12_+_21679220 | 0.18 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr6_+_151561506 | 0.17 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr9_+_44867571 | 0.17 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr7_+_26591441 | 0.16 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr6_+_155334780 | 0.16 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_+_151561085 | 0.15 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr17_-_46608272 | 0.15 |
ENST00000577092.1
ENST00000239174.6 |
HOXB1
|
homeobox B1 |
| chr17_-_46623441 | 0.14 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr15_-_64673630 | 0.13 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr2_+_12857043 | 0.13 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr11_-_2162162 | 0.12 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr1_-_205391178 | 0.12 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr8_+_143530791 | 0.11 |
ENST00000517894.1
|
BAI1
|
brain-specific angiogenesis inhibitor 1 |
| chr3_+_239652 | 0.11 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr8_-_67874805 | 0.10 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr8_+_23386305 | 0.10 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr19_-_45735138 | 0.09 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr8_-_79717750 | 0.08 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr5_-_125930929 | 0.08 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr9_-_26946981 | 0.08 |
ENST00000523212.1
|
PLAA
|
phospholipase A2-activating protein |
| chr15_-_64673665 | 0.08 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr1_-_217804377 | 0.07 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr2_+_68961905 | 0.07 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_+_7023491 | 0.07 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr14_-_54425475 | 0.07 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr3_-_156877997 | 0.07 |
ENST00000295926.3
|
CCNL1
|
cyclin L1 |
| chr2_+_68961934 | 0.07 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr17_-_42277203 | 0.07 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr8_+_77596014 | 0.07 |
ENST00000523885.1
|
ZFHX4
|
zinc finger homeobox 4 |
| chr1_-_43833628 | 0.07 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr8_+_23386557 | 0.06 |
ENST00000523930.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr5_+_154181816 | 0.06 |
ENST00000518677.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr7_+_86273218 | 0.06 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr9_+_113431029 | 0.06 |
ENST00000189978.5
ENST00000374448.4 ENST00000374440.3 |
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr2_+_68962014 | 0.06 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr9_-_26947220 | 0.06 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
| chr1_+_68150744 | 0.06 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr6_+_32006042 | 0.06 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr2_+_12857015 | 0.06 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr19_+_37997812 | 0.06 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr8_-_128231299 | 0.06 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr19_+_13051206 | 0.06 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr5_+_167181917 | 0.05 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr9_-_15250114 | 0.05 |
ENST00000507993.1
|
TTC39B
|
tetratricopeptide repeat domain 39B |
| chrX_+_41583408 | 0.05 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr17_-_1588101 | 0.05 |
ENST00000577001.1
ENST00000572621.1 ENST00000304992.6 |
PRPF8
|
pre-mRNA processing factor 8 |
| chr12_+_7022909 | 0.05 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr11_-_93583697 | 0.05 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr9_+_140135665 | 0.05 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chrX_-_13835398 | 0.05 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr2_+_135011731 | 0.05 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr5_+_147582387 | 0.05 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr16_-_49890016 | 0.05 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr19_+_52074502 | 0.05 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr11_-_122930121 | 0.05 |
ENST00000524552.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr17_-_27418537 | 0.05 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr11_+_57365150 | 0.05 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr3_-_150264272 | 0.04 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr3_+_159481988 | 0.04 |
ENST00000472451.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr6_-_159065741 | 0.04 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr10_-_98945515 | 0.04 |
ENST00000371070.4
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr9_-_26947453 | 0.04 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr17_+_37824217 | 0.04 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr15_-_74501360 | 0.04 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr2_-_242089677 | 0.04 |
ENST00000405260.1
|
PASK
|
PAS domain containing serine/threonine kinase |
| chr1_-_217250231 | 0.04 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr11_-_122929699 | 0.04 |
ENST00000526686.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr13_+_52586517 | 0.04 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr11_+_133902547 | 0.03 |
ENST00000529070.1
|
RP11-713P17.3
|
RP11-713P17.3 |
| chr13_+_36050881 | 0.03 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr17_+_37824411 | 0.03 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr15_-_38852251 | 0.03 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr12_-_28125638 | 0.03 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr1_-_109584608 | 0.03 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr1_-_109584768 | 0.03 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr10_-_98945677 | 0.03 |
ENST00000266058.4
ENST00000371041.3 |
SLIT1
|
slit homolog 1 (Drosophila) |
| chr12_-_28124903 | 0.03 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr17_-_42276574 | 0.03 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr1_-_109584716 | 0.03 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr7_-_78400364 | 0.03 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_-_24733444 | 0.02 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chr2_+_114647504 | 0.02 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr3_+_191046810 | 0.02 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr9_+_113431059 | 0.02 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr1_+_52082751 | 0.02 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr17_+_79495397 | 0.02 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr7_+_20370746 | 0.02 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr12_-_86650077 | 0.02 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_+_243419306 | 0.02 |
ENST00000355875.4
ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8
|
serologically defined colon cancer antigen 8 |
| chrX_-_30993201 | 0.02 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr12_+_10658489 | 0.02 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr2_+_54684327 | 0.02 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr5_+_98109322 | 0.02 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr2_+_242089833 | 0.02 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr4_-_90757364 | 0.02 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr10_+_124320156 | 0.02 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr3_+_150264555 | 0.02 |
ENST00000406576.3
ENST00000482093.1 ENST00000273435.5 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr5_+_133861790 | 0.02 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr11_+_68228186 | 0.02 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
| chr7_+_39017504 | 0.02 |
ENST00000403058.1
|
POU6F2
|
POU class 6 homeobox 2 |
| chr12_-_114843889 | 0.02 |
ENST00000405440.2
|
TBX5
|
T-box 5 |
| chr3_+_150264458 | 0.02 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr1_+_36621174 | 0.02 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr15_+_81591757 | 0.02 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr20_+_30467600 | 0.02 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr17_+_43239191 | 0.01 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chrX_-_110655306 | 0.01 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr3_+_189507460 | 0.01 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr11_-_126870655 | 0.01 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr11_-_128712362 | 0.01 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr15_-_74501310 | 0.01 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr2_+_176995011 | 0.01 |
ENST00000548663.1
ENST00000450510.2 |
HOXD8
|
homeobox D8 |
| chr19_-_51512804 | 0.01 |
ENST00000594211.1
ENST00000376832.4 |
KLK9
|
kallikrein-related peptidase 9 |
| chr21_-_43430440 | 0.01 |
ENST00000398505.3
ENST00000310826.5 ENST00000449949.1 ENST00000398499.1 ENST00000398497.2 ENST00000398511.3 |
ZBTB21
|
zinc finger and BTB domain containing 21 |
| chr10_-_98945264 | 0.01 |
ENST00000314867.5
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr21_+_19617140 | 0.01 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr20_+_21284416 | 0.01 |
ENST00000539513.1
|
XRN2
|
5'-3' exoribonuclease 2 |
| chr9_-_73477826 | 0.01 |
ENST00000396285.1
ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chrX_+_95939711 | 0.01 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr13_-_36050819 | 0.01 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr17_+_39975544 | 0.01 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr12_+_56414795 | 0.01 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr3_-_98619999 | 0.01 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr2_+_26624775 | 0.01 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr5_-_125930877 | 0.01 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr6_+_32006159 | 0.01 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr17_+_39975455 | 0.01 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr6_-_112575838 | 0.01 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr12_+_56414851 | 0.01 |
ENST00000547167.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr10_+_124320195 | 0.01 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr20_+_21283941 | 0.01 |
ENST00000377191.3
ENST00000430571.2 |
XRN2
|
5'-3' exoribonuclease 2 |
| chr2_-_145278475 | 0.01 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_64029879 | 0.01 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr8_-_22550691 | 0.01 |
ENST00000519492.1
|
EGR3
|
early growth response 3 |
| chr4_-_90756769 | 0.01 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr3_+_189507432 | 0.00 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr1_-_92351666 | 0.00 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chrX_+_99899180 | 0.00 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr2_-_197226875 | 0.00 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_+_121493717 | 0.00 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr4_+_129732467 | 0.00 |
ENST00000413543.2
|
PHF17
|
jade family PHD finger 1 |
| chr4_+_77356248 | 0.00 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr12_+_101988774 | 0.00 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr10_+_24755416 | 0.00 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr12_+_101988627 | 0.00 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chrX_-_110655391 | 0.00 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr7_-_86595190 | 0.00 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr6_+_33387868 | 0.00 |
ENST00000418600.2
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA2_HOXB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.1 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:2000006 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |