Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
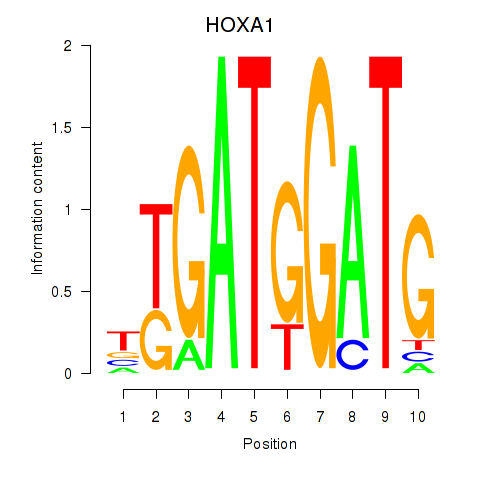
Results for HOXA1
Z-value: 0.37
Transcription factors associated with HOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA1
|
ENSG00000105991.7 | homeobox A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA1 | hg19_v2_chr7_-_27135591_27135658 | -0.02 | 9.8e-01 | Click! |
Activity profile of HOXA1 motif
Sorted Z-values of HOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_41968007 | 0.68 |
ENST00000460790.1
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr22_-_30642728 | 0.35 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr20_-_36794902 | 0.24 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr20_-_36794938 | 0.24 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr9_-_5304432 | 0.22 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr3_+_125985620 | 0.18 |
ENST00000511512.1
ENST00000512435.1 |
RP11-71E19.1
|
RP11-71E19.1 |
| chr12_+_78359999 | 0.17 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr15_-_74501310 | 0.17 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr19_+_5681011 | 0.16 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr1_-_152539248 | 0.15 |
ENST00000368789.1
|
LCE3E
|
late cornified envelope 3E |
| chr17_-_57229155 | 0.15 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_-_38807199 | 0.14 |
ENST00000508364.1
|
TLR1
|
toll-like receptor 1 |
| chr3_-_129375556 | 0.14 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr14_+_102196739 | 0.13 |
ENST00000556973.1
|
RP11-796G6.2
|
Uncharacterized protein |
| chr19_-_38746979 | 0.13 |
ENST00000591291.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr22_-_22641542 | 0.13 |
ENST00000606654.1
|
LL22NC03-2H8.5
|
LL22NC03-2H8.5 |
| chr2_+_24272576 | 0.10 |
ENST00000380986.4
ENST00000452109.1 |
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr8_-_42360015 | 0.10 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr5_+_162864575 | 0.10 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr8_-_91095099 | 0.10 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr8_+_104152922 | 0.10 |
ENST00000309982.5
ENST00000438105.2 ENST00000297574.6 |
BAALC
|
brain and acute leukemia, cytoplasmic |
| chr5_+_159656437 | 0.09 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr10_+_60272814 | 0.09 |
ENST00000373886.3
|
BICC1
|
bicaudal C homolog 1 (Drosophila) |
| chr8_+_40010989 | 0.09 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr12_-_86650077 | 0.09 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr16_-_55867146 | 0.09 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr12_-_106477805 | 0.08 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr9_+_33795533 | 0.08 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr19_-_18654828 | 0.08 |
ENST00000609656.1
ENST00000597611.3 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr17_-_40835076 | 0.08 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr11_+_61248583 | 0.08 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr3_-_145878954 | 0.08 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr14_+_96949319 | 0.08 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr17_-_42276574 | 0.07 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr1_+_65730385 | 0.07 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr19_-_40732594 | 0.07 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr6_+_32006042 | 0.07 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr20_+_60174827 | 0.07 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr2_+_24272543 | 0.07 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr3_-_98619999 | 0.07 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr16_-_11617444 | 0.07 |
ENST00000598234.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr17_-_15502111 | 0.07 |
ENST00000354433.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr1_+_17634689 | 0.07 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr11_+_2482661 | 0.07 |
ENST00000335475.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr5_+_36152091 | 0.07 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr2_-_75788038 | 0.07 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr2_-_239148599 | 0.07 |
ENST00000409182.1
ENST00000409002.3 ENST00000450098.1 ENST00000409356.1 ENST00000409160.3 ENST00000409574.1 ENST00000272937.5 |
HES6
|
hes family bHLH transcription factor 6 |
| chr12_-_95510743 | 0.07 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr7_-_994302 | 0.06 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr20_+_44036620 | 0.06 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr13_+_28519343 | 0.06 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr1_+_36621174 | 0.06 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr17_-_15501932 | 0.06 |
ENST00000583965.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr3_-_161090660 | 0.06 |
ENST00000359175.4
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr3_+_127872265 | 0.06 |
ENST00000254730.6
ENST00000483457.1 |
EEFSEC
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr12_+_48577366 | 0.06 |
ENST00000316554.3
|
C12orf68
|
chromosome 12 open reading frame 68 |
| chr1_+_165864800 | 0.06 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr21_-_32119551 | 0.06 |
ENST00000333892.2
|
KRTAP21-2
|
keratin associated protein 21-2 |
| chr12_+_8276433 | 0.06 |
ENST00000345999.3
ENST00000352620.3 ENST00000360500.3 |
CLEC4A
|
C-type lectin domain family 4, member A |
| chr8_-_37411648 | 0.05 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr1_+_45140360 | 0.05 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr16_+_30080857 | 0.05 |
ENST00000565355.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr22_-_41677625 | 0.05 |
ENST00000452543.1
ENST00000418067.1 |
RANGAP1
|
Ran GTPase activating protein 1 |
| chr9_-_89562104 | 0.05 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chrX_+_150148976 | 0.05 |
ENST00000419110.1
|
HMGB3
|
high mobility group box 3 |
| chr5_-_141338627 | 0.05 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr2_+_121011027 | 0.05 |
ENST00000449649.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr20_+_44036900 | 0.05 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_+_66247478 | 0.05 |
ENST00000531863.1
ENST00000532677.1 |
DPP3
|
dipeptidyl-peptidase 3 |
| chr12_-_110906027 | 0.05 |
ENST00000537466.2
ENST00000550974.1 ENST00000228827.3 |
GPN3
|
GPN-loop GTPase 3 |
| chr11_-_93583697 | 0.05 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr19_+_5681153 | 0.05 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr1_-_153513170 | 0.05 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr19_-_45735138 | 0.05 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr1_-_32384693 | 0.05 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr1_-_217804377 | 0.05 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr1_+_183155373 | 0.05 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr3_-_146262488 | 0.05 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr15_-_42343388 | 0.05 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr3_+_44916098 | 0.05 |
ENST00000296125.4
|
TGM4
|
transglutaminase 4 |
| chr4_+_177241094 | 0.04 |
ENST00000503362.1
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr9_-_128246769 | 0.04 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr19_+_19144384 | 0.04 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr13_+_113863858 | 0.04 |
ENST00000375440.4
|
CUL4A
|
cullin 4A |
| chr17_-_8059638 | 0.04 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr20_-_45976816 | 0.04 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr7_-_2354099 | 0.04 |
ENST00000222990.3
|
SNX8
|
sorting nexin 8 |
| chr6_-_44233361 | 0.04 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr22_-_22307199 | 0.04 |
ENST00000397495.4
ENST00000263212.5 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr10_+_90562705 | 0.04 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chrX_+_57618269 | 0.04 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr10_-_101380121 | 0.04 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr10_-_5638048 | 0.04 |
ENST00000478294.1
|
RP13-463N16.6
|
RP13-463N16.6 |
| chr10_-_106240032 | 0.04 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr11_-_111649015 | 0.04 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_-_179322436 | 0.04 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr5_+_133861339 | 0.04 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr11_+_36317830 | 0.03 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr14_+_65453432 | 0.03 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr11_-_2162162 | 0.03 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_-_150264272 | 0.03 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr17_-_72619869 | 0.03 |
ENST00000392619.1
ENST00000426295.2 |
CD300E
|
CD300e molecule |
| chr15_+_67418047 | 0.03 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr9_+_10613163 | 0.03 |
ENST00000429581.2
|
RP11-87N24.2
|
RP11-87N24.2 |
| chr5_+_150040403 | 0.03 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr17_+_7758374 | 0.03 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr19_+_33072373 | 0.03 |
ENST00000586035.1
|
PDCD5
|
programmed cell death 5 |
| chr10_+_89264625 | 0.03 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
| chr17_-_1395954 | 0.03 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr9_-_116861337 | 0.03 |
ENST00000374118.3
|
KIF12
|
kinesin family member 12 |
| chr12_+_123459127 | 0.03 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr11_+_60739249 | 0.03 |
ENST00000542157.1
ENST00000433107.2 ENST00000452451.2 ENST00000352009.5 |
CD6
|
CD6 molecule |
| chr2_+_264869 | 0.03 |
ENST00000272067.6
ENST00000272065.5 ENST00000407983.3 |
ACP1
|
acid phosphatase 1, soluble |
| chr19_-_42192096 | 0.03 |
ENST00000602225.1
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr13_-_46425865 | 0.03 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr16_+_28303804 | 0.03 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr20_-_62168714 | 0.03 |
ENST00000542869.1
|
PTK6
|
protein tyrosine kinase 6 |
| chr12_-_89919965 | 0.03 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr2_-_175711978 | 0.03 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr7_+_100136811 | 0.03 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr5_+_166711804 | 0.03 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr14_-_106692191 | 0.03 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr20_-_3154162 | 0.03 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr10_-_9801179 | 0.03 |
ENST00000419836.1
|
RP5-1051H14.2
|
RP5-1051H14.2 |
| chr7_+_142448053 | 0.03 |
ENST00000422143.2
|
TRBV29-1
|
T cell receptor beta variable 29-1 |
| chr18_+_22006646 | 0.03 |
ENST00000585067.1
ENST00000578221.1 |
IMPACT
|
impact RWD domain protein |
| chr7_+_30174668 | 0.03 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_179322416 | 0.03 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr14_-_23504432 | 0.03 |
ENST00000425762.2
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr16_-_21431078 | 0.03 |
ENST00000458643.2
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr16_+_29674540 | 0.03 |
ENST00000436527.1
ENST00000360121.3 ENST00000449759.1 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr9_-_97402531 | 0.03 |
ENST00000415431.1
|
FBP1
|
fructose-1,6-bisphosphatase 1 |
| chr19_-_5680891 | 0.03 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr9_+_132962843 | 0.03 |
ENST00000458469.1
|
NCS1
|
neuronal calcium sensor 1 |
| chr11_+_63706444 | 0.03 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr14_-_23504337 | 0.03 |
ENST00000361611.6
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr5_-_137071689 | 0.03 |
ENST00000505853.1
|
KLHL3
|
kelch-like family member 3 |
| chr10_-_126849068 | 0.03 |
ENST00000494626.2
ENST00000337195.5 |
CTBP2
|
C-terminal binding protein 2 |
| chr15_-_60695071 | 0.03 |
ENST00000557904.1
|
ANXA2
|
annexin A2 |
| chr8_-_66546373 | 0.03 |
ENST00000518908.1
ENST00000458464.2 ENST00000519352.1 |
ARMC1
|
armadillo repeat containing 1 |
| chr3_-_112564797 | 0.03 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr6_-_36725157 | 0.03 |
ENST00000393189.2
|
CPNE5
|
copine V |
| chr1_+_45140400 | 0.02 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr19_+_38880695 | 0.02 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr11_-_111649074 | 0.02 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chrX_+_139791917 | 0.02 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr3_+_98216448 | 0.02 |
ENST00000427338.1
|
OR5K2
|
olfactory receptor, family 5, subfamily K, member 2 |
| chr10_+_99349450 | 0.02 |
ENST00000370640.3
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr1_+_26872324 | 0.02 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr17_+_60447579 | 0.02 |
ENST00000450662.2
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr7_+_151791095 | 0.02 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr14_+_24779340 | 0.02 |
ENST00000533293.1
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr9_-_10612703 | 0.02 |
ENST00000463477.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr12_+_12878829 | 0.02 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr3_-_54962100 | 0.02 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr7_+_20370746 | 0.02 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chrX_+_48455866 | 0.02 |
ENST00000376729.5
ENST00000218056.5 |
WDR13
|
WD repeat domain 13 |
| chr14_-_101053739 | 0.02 |
ENST00000554140.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr9_+_116037922 | 0.02 |
ENST00000374198.4
|
PRPF4
|
pre-mRNA processing factor 4 |
| chr5_+_125758865 | 0.02 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr12_-_76817036 | 0.02 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr12_-_85306562 | 0.02 |
ENST00000551612.1
ENST00000450363.3 ENST00000552192.1 |
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr17_-_26989136 | 0.02 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr16_+_28986134 | 0.02 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr6_+_73331520 | 0.02 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr21_-_46340884 | 0.02 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr20_+_36012051 | 0.02 |
ENST00000373567.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr17_+_4046964 | 0.02 |
ENST00000573984.1
|
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr19_-_15544099 | 0.02 |
ENST00000599910.2
|
WIZ
|
widely interspaced zinc finger motifs |
| chr2_-_170681375 | 0.02 |
ENST00000410097.1
ENST00000308099.3 ENST00000409837.1 ENST00000538491.1 ENST00000260953.5 ENST00000409965.1 ENST00000392640.2 |
METTL5
|
methyltransferase like 5 |
| chr1_+_107682629 | 0.02 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr6_+_151561085 | 0.02 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr12_-_85306594 | 0.02 |
ENST00000266682.5
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr10_-_134756030 | 0.02 |
ENST00000368586.5
ENST00000368582.2 |
TTC40
|
tetratricopeptide repeat domain 40 |
| chr22_-_19419205 | 0.02 |
ENST00000340170.4
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr17_+_7328925 | 0.02 |
ENST00000333870.3
ENST00000574034.1 |
C17orf74
|
chromosome 17 open reading frame 74 |
| chr7_-_115670804 | 0.02 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr1_-_182369751 | 0.02 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr16_-_20566616 | 0.02 |
ENST00000569163.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr1_+_205473720 | 0.02 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr7_+_151791037 | 0.02 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr9_+_35829208 | 0.02 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr13_+_102104952 | 0.02 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr5_+_167181917 | 0.02 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr14_+_23709555 | 0.02 |
ENST00000430154.2
|
C14orf164
|
chromosome 14 open reading frame 164 |
| chr10_-_81932771 | 0.02 |
ENST00000437799.1
|
ANXA11
|
annexin A11 |
| chr15_-_34635314 | 0.02 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr13_+_102104980 | 0.02 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr7_+_79765071 | 0.02 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr4_+_113970772 | 0.02 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr2_-_190044480 | 0.02 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr9_-_130679257 | 0.02 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr7_+_94139105 | 0.02 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr6_+_151561506 | 0.02 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_+_27255806 | 0.02 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr15_-_81666546 | 0.02 |
ENST00000558726.1
ENST00000359440.5 |
TMC3
|
transmembrane channel-like 3 |
| chr8_+_68864330 | 0.02 |
ENST00000288368.4
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_2005425 | 0.02 |
ENST00000461106.2
|
PRKCZ
|
protein kinase C, zeta |
| chr22_+_39101728 | 0.02 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr8_-_119124045 | 0.02 |
ENST00000378204.2
|
EXT1
|
exostosin glycosyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.2 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.0 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |