Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
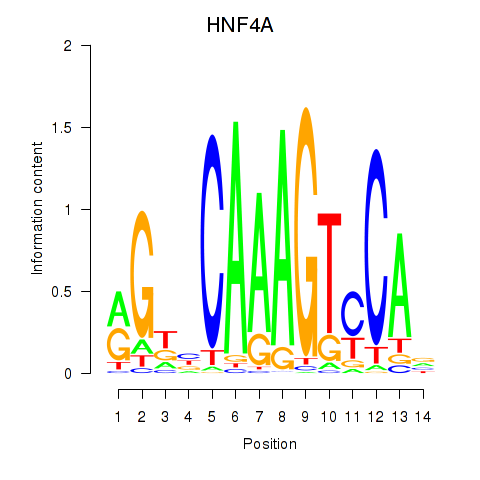
Results for HNF4A
Z-value: 0.82
Transcription factors associated with HNF4A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4A
|
ENSG00000101076.12 | hepatocyte nuclear factor 4 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4A | hg19_v2_chr20_+_43029911_43029941 | -0.20 | 7.0e-01 | Click! |
Activity profile of HNF4A motif
Sorted Z-values of HNF4A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_42811986 | 0.67 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr2_-_222436988 | 0.59 |
ENST00000409854.1
ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4
|
EPH receptor A4 |
| chr5_-_42812143 | 0.56 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_-_177939348 | 0.45 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr19_+_45116921 | 0.41 |
ENST00000402988.1
|
IGSF23
|
immunoglobulin superfamily, member 23 |
| chr4_+_113152978 | 0.41 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr1_+_23695680 | 0.40 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr1_-_43751230 | 0.39 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr14_-_76447494 | 0.38 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr2_-_222437049 | 0.35 |
ENST00000541600.1
|
EPHA4
|
EPH receptor A4 |
| chr3_-_158390282 | 0.33 |
ENST00000264265.3
|
LXN
|
latexin |
| chr17_+_73606766 | 0.33 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chr2_+_97454321 | 0.32 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr11_-_73720276 | 0.32 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr17_+_4692230 | 0.31 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr11_+_1856034 | 0.30 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr12_-_95010147 | 0.29 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr2_-_74648702 | 0.27 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr6_+_31950150 | 0.27 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr2_+_14772810 | 0.24 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr1_+_78511586 | 0.24 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr1_+_94883991 | 0.24 |
ENST00000370214.4
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr11_+_27076764 | 0.23 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_+_6508100 | 0.23 |
ENST00000461727.1
|
ESPN
|
espin |
| chr1_+_100435315 | 0.22 |
ENST00000370155.3
ENST00000465289.1 |
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr2_-_74645669 | 0.21 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr11_+_1855645 | 0.21 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr15_-_100273544 | 0.21 |
ENST00000409796.1
ENST00000545021.1 ENST00000344791.2 ENST00000332728.4 ENST00000450512.1 |
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr12_+_121163538 | 0.21 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr1_-_161207875 | 0.20 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_94883931 | 0.20 |
ENST00000394233.2
ENST00000454898.2 ENST00000536817.1 |
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr10_-_116444371 | 0.20 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr17_+_1646130 | 0.20 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr1_+_100435535 | 0.19 |
ENST00000427993.2
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr1_-_161208013 | 0.18 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr12_+_56114151 | 0.18 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr6_-_30080876 | 0.18 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr6_+_30882108 | 0.18 |
ENST00000541562.1
ENST00000421263.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr12_-_7125770 | 0.17 |
ENST00000261407.4
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr10_-_96829246 | 0.17 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr3_+_37284824 | 0.17 |
ENST00000431105.1
|
GOLGA4
|
golgin A4 |
| chr22_+_27068704 | 0.17 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr8_+_70404996 | 0.16 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr5_-_79950371 | 0.16 |
ENST00000511032.1
ENST00000504396.1 ENST00000505337.1 |
DHFR
|
dihydrofolate reductase |
| chr19_-_49339080 | 0.16 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr20_+_36661910 | 0.16 |
ENST00000373433.4
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr20_-_36156264 | 0.16 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr12_-_118810688 | 0.16 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr4_-_10023095 | 0.15 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr3_+_37284668 | 0.15 |
ENST00000361924.2
ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4
|
golgin A4 |
| chr1_-_211848899 | 0.15 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr1_-_23694794 | 0.15 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr14_+_101295948 | 0.15 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr12_-_53594227 | 0.15 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr18_-_61034743 | 0.15 |
ENST00000406396.3
|
KDSR
|
3-ketodihydrosphingosine reductase |
| chr16_-_3350614 | 0.15 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr13_+_50656307 | 0.15 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr17_-_28618867 | 0.15 |
ENST00000394819.3
ENST00000577623.1 |
BLMH
|
bleomycin hydrolase |
| chr12_+_56114189 | 0.14 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr12_+_121163602 | 0.14 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr2_+_159313452 | 0.14 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr3_-_194072019 | 0.13 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr2_+_191792376 | 0.13 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr1_-_177939041 | 0.13 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr4_+_159593271 | 0.13 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr9_-_27005686 | 0.13 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr2_-_219433014 | 0.13 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr19_-_42758040 | 0.12 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr4_-_120243545 | 0.12 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr5_+_14664762 | 0.12 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr15_-_41120896 | 0.12 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr18_-_54318353 | 0.12 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr11_-_63536113 | 0.12 |
ENST00000433688.1
ENST00000546282.2 |
C11orf95
RP11-466C23.4
|
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chr17_-_36105009 | 0.12 |
ENST00000560016.1
ENST00000427275.2 ENST00000561193.1 |
HNF1B
|
HNF1 homeobox B |
| chr12_+_67663056 | 0.12 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr3_-_125775629 | 0.12 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr6_+_80341000 | 0.12 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr1_-_161207986 | 0.12 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_+_63898275 | 0.12 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr12_-_95009837 | 0.11 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr5_+_133842243 | 0.11 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr17_-_7082861 | 0.11 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr14_-_76447336 | 0.11 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor, beta 3 |
| chr16_+_21244986 | 0.11 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr2_+_219433588 | 0.11 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr4_+_113152881 | 0.11 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr19_+_10197463 | 0.10 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr5_-_16509101 | 0.10 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr6_-_30080863 | 0.10 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chrX_-_77041685 | 0.10 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr2_-_180871780 | 0.10 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr10_-_21186144 | 0.10 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr20_+_62694461 | 0.10 |
ENST00000343484.5
ENST00000395053.3 |
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr3_+_63897605 | 0.09 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr9_-_136605042 | 0.09 |
ENST00000371872.4
ENST00000298628.5 ENST00000422262.2 |
SARDH
|
sarcosine dehydrogenase |
| chr3_-_125313934 | 0.09 |
ENST00000296220.5
|
OSBPL11
|
oxysterol binding protein-like 11 |
| chr19_+_4639514 | 0.09 |
ENST00000327473.4
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr1_-_161207953 | 0.09 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr6_+_30881982 | 0.09 |
ENST00000321897.5
ENST00000416670.2 ENST00000542001.1 ENST00000428017.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr19_+_7660716 | 0.09 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr8_+_96037255 | 0.09 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr16_+_66600294 | 0.09 |
ENST00000535705.1
ENST00000332695.7 ENST00000336328.6 ENST00000528324.1 ENST00000531885.1 ENST00000529506.1 ENST00000457188.2 ENST00000533666.1 ENST00000533953.1 ENST00000379500.2 ENST00000328020.6 |
CMTM1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr14_-_23446003 | 0.09 |
ENST00000553911.1
|
AJUBA
|
ajuba LIM protein |
| chr16_-_88752889 | 0.08 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr10_+_102106829 | 0.08 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr12_-_54121261 | 0.08 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr15_+_81489213 | 0.08 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr4_+_144312659 | 0.08 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr2_-_43019698 | 0.08 |
ENST00000431905.1
ENST00000294973.6 |
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr2_+_48796120 | 0.08 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr17_-_38256973 | 0.07 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr17_-_27503770 | 0.07 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr20_+_44486246 | 0.07 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr10_+_96698406 | 0.07 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr9_-_15307200 | 0.07 |
ENST00000506891.1
ENST00000541445.1 ENST00000512701.2 ENST00000380850.4 ENST00000297615.5 ENST00000355694.2 |
TTC39B
|
tetratricopeptide repeat domain 39B |
| chr15_+_101142722 | 0.07 |
ENST00000332783.7
ENST00000558747.1 ENST00000343276.4 |
ASB7
|
ankyrin repeat and SOCS box containing 7 |
| chr15_+_50716576 | 0.07 |
ENST00000560297.1
ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8
|
ubiquitin specific peptidase 8 |
| chr3_-_141868293 | 0.07 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_-_54121212 | 0.07 |
ENST00000548263.1
ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chrX_+_65382433 | 0.06 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr6_+_31691121 | 0.06 |
ENST00000480039.1
ENST00000375810.4 ENST00000375805.2 ENST00000375809.3 ENST00000375804.2 ENST00000375814.3 ENST00000375806.2 |
C6orf25
|
chromosome 6 open reading frame 25 |
| chr7_+_100663353 | 0.06 |
ENST00000306151.4
|
MUC17
|
mucin 17, cell surface associated |
| chrX_+_65382381 | 0.06 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr11_-_64660916 | 0.06 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr18_+_34124507 | 0.06 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr17_+_41857793 | 0.06 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr6_+_31949801 | 0.06 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr2_-_39348137 | 0.06 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr4_-_120225600 | 0.06 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr19_+_39881951 | 0.06 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr1_-_11863171 | 0.06 |
ENST00000376592.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr2_-_21266816 | 0.05 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr14_+_23790655 | 0.05 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr17_-_7082668 | 0.05 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr22_+_27068766 | 0.05 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr1_+_180601139 | 0.05 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr1_-_15911510 | 0.05 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr6_+_31982539 | 0.05 |
ENST00000435363.2
ENST00000425700.2 |
C4B
|
complement component 4B (Chido blood group) |
| chr1_+_113217309 | 0.05 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr15_-_101142362 | 0.05 |
ENST00000559577.1
ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS
|
lines homolog (Drosophila) |
| chr15_+_63340775 | 0.05 |
ENST00000559281.1
ENST00000317516.7 |
TPM1
|
tropomyosin 1 (alpha) |
| chr15_+_63340858 | 0.05 |
ENST00000560615.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr12_-_121973974 | 0.05 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr5_+_136070614 | 0.05 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr19_+_1205740 | 0.05 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr8_-_98290087 | 0.05 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr14_+_23352374 | 0.05 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr12_-_6665200 | 0.05 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr11_-_64163297 | 0.05 |
ENST00000457725.1
|
AP003774.6
|
AP003774.6 |
| chr10_+_81065975 | 0.04 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr19_-_12886327 | 0.04 |
ENST00000397668.3
ENST00000587178.1 ENST00000264827.5 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr13_+_52586517 | 0.04 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr17_+_4853442 | 0.04 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr12_-_46384334 | 0.04 |
ENST00000369367.3
ENST00000266589.6 ENST00000395453.2 ENST00000395454.2 |
SCAF11
|
SR-related CTD-associated factor 11 |
| chr12_+_121416489 | 0.04 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr16_+_19429018 | 0.04 |
ENST00000542583.2
|
TMC5
|
transmembrane channel-like 5 |
| chr19_+_10196981 | 0.04 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_-_71750865 | 0.04 |
ENST00000544129.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr7_+_33765593 | 0.04 |
ENST00000311067.3
|
RP11-89N17.1
|
HCG1643653; Uncharacterized protein |
| chr14_+_39735411 | 0.04 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr12_+_53497263 | 0.04 |
ENST00000551896.1
ENST00000301466.3 |
SOAT2
|
sterol O-acyltransferase 2 |
| chr14_+_23790690 | 0.04 |
ENST00000556821.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr6_+_43028182 | 0.04 |
ENST00000394058.1
|
KLC4
|
kinesin light chain 4 |
| chr14_+_77647966 | 0.04 |
ENST00000554766.1
|
TMEM63C
|
transmembrane protein 63C |
| chr3_-_120400960 | 0.04 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr5_-_32444828 | 0.03 |
ENST00000265069.8
|
ZFR
|
zinc finger RNA binding protein |
| chr15_+_63340734 | 0.03 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr11_+_119056178 | 0.03 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr10_-_103815874 | 0.03 |
ENST00000370033.4
ENST00000311122.5 |
C10orf76
|
chromosome 10 open reading frame 76 |
| chr19_-_35625765 | 0.03 |
ENST00000591633.1
|
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr1_-_43751276 | 0.03 |
ENST00000423420.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr8_+_104831554 | 0.03 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_+_70320413 | 0.03 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr11_-_71781096 | 0.03 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr1_+_113217073 | 0.03 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_-_41131326 | 0.03 |
ENST00000372684.3
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr12_+_121416437 | 0.03 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr6_+_32146131 | 0.03 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr2_+_219135115 | 0.02 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr17_-_2614927 | 0.02 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr1_+_241695670 | 0.02 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_-_48867171 | 0.02 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr7_+_143080063 | 0.02 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr19_+_46806856 | 0.02 |
ENST00000300862.3
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr1_-_29508321 | 0.02 |
ENST00000546138.1
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr9_-_35685452 | 0.02 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr17_-_44896047 | 0.02 |
ENST00000225512.5
|
WNT3
|
wingless-type MMTV integration site family, member 3 |
| chr22_+_19950060 | 0.02 |
ENST00000449653.1
|
COMT
|
catechol-O-methyltransferase |
| chr6_-_33385823 | 0.02 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr20_-_36156293 | 0.02 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chr2_-_21266935 | 0.01 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr1_+_241695424 | 0.01 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr12_-_39837192 | 0.01 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr3_+_9745510 | 0.01 |
ENST00000383831.3
|
CPNE9
|
copine family member IX |
| chr6_+_147527103 | 0.01 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr3_+_123813509 | 0.01 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr1_+_113217345 | 0.01 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_-_26197744 | 0.01 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr4_+_109541740 | 0.01 |
ENST00000394665.1
|
RPL34
|
ribosomal protein L34 |
| chr1_-_151138323 | 0.01 |
ENST00000368908.5
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 0.5 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.2 | 0.5 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.1 | 0.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.4 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.2 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.2 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.0 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.0 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |