Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
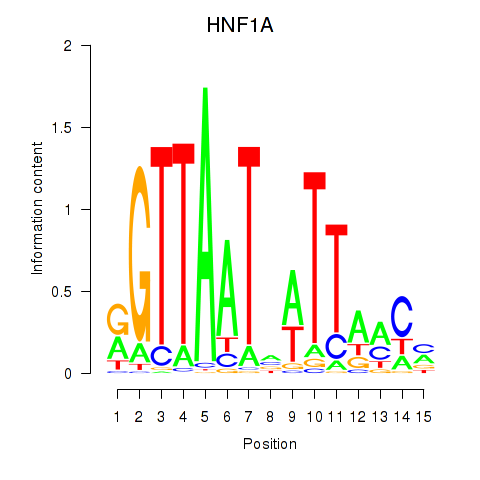
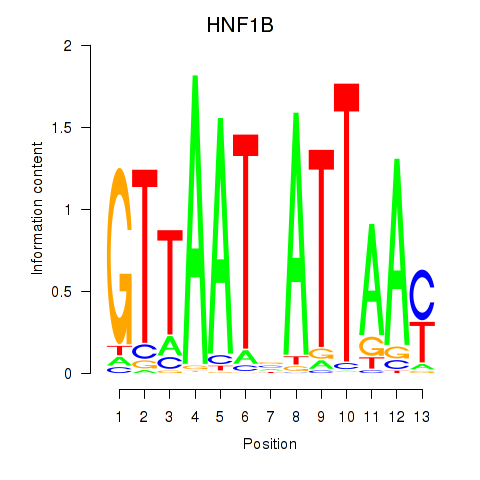
Results for HNF1A_HNF1B
Z-value: 0.85
Transcription factors associated with HNF1A_HNF1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF1A
|
ENSG00000135100.13 | HNF1 homeobox A |
|
HNF1B
|
ENSG00000108753.8 | HNF1 homeobox B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF1B | hg19_v2_chr17_-_36105009_36105060 | 0.71 | 1.1e-01 | Click! |
| HNF1A | hg19_v2_chr12_+_121416489_121416552 | 0.57 | 2.4e-01 | Click! |
Activity profile of HNF1A_HNF1B motif
Sorted Z-values of HNF1A_HNF1B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_30899924 | 0.95 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr4_-_110723335 | 0.57 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr4_-_110723134 | 0.51 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr3_-_161089289 | 0.46 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr4_-_110723194 | 0.46 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr4_+_74301880 | 0.46 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr16_-_69385681 | 0.36 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr2_-_47572105 | 0.35 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr11_-_117170403 | 0.30 |
ENST00000504995.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr2_-_47572207 | 0.27 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr5_+_176513868 | 0.25 |
ENST00000292408.4
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr14_+_39703112 | 0.24 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr22_-_32767017 | 0.24 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr5_+_125706998 | 0.24 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr10_+_101542462 | 0.24 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr14_-_57960545 | 0.24 |
ENST00000526336.1
ENST00000216445.3 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr14_+_65170820 | 0.23 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr4_+_39408470 | 0.23 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr11_-_16419067 | 0.23 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_204183071 | 0.23 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr14_+_38033252 | 0.23 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr19_+_49467232 | 0.22 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr11_-_107729287 | 0.21 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr19_+_50016610 | 0.21 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr20_+_42187682 | 0.20 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr12_+_40787194 | 0.20 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr7_-_150020814 | 0.19 |
ENST00000477871.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr12_+_25205568 | 0.19 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr4_-_1166954 | 0.19 |
ENST00000514490.1
ENST00000431380.1 ENST00000503765.1 |
SPON2
|
spondin 2, extracellular matrix protein |
| chr16_+_22524379 | 0.18 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr16_+_21244986 | 0.17 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr4_-_987164 | 0.17 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr12_+_25205666 | 0.17 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr4_+_8201091 | 0.16 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr3_-_126327398 | 0.16 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr20_+_42187608 | 0.16 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr6_+_131571535 | 0.16 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr1_+_162351503 | 0.16 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr12_+_25205628 | 0.16 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr5_+_176513895 | 0.15 |
ENST00000503708.1
ENST00000393648.2 ENST00000514472.1 ENST00000502906.1 ENST00000292410.3 ENST00000510911.1 |
FGFR4
|
fibroblast growth factor receptor 4 |
| chr2_+_234601512 | 0.15 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr16_+_20775358 | 0.15 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr2_+_27719697 | 0.15 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr19_+_50016411 | 0.15 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_+_560956 | 0.14 |
ENST00000397582.3
ENST00000344375.4 ENST00000397583.3 |
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr12_+_93096619 | 0.14 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr10_+_135340859 | 0.14 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr1_+_18807424 | 0.14 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr3_-_137834436 | 0.14 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr10_+_5135981 | 0.13 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr7_-_44580861 | 0.13 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr16_+_82068873 | 0.12 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr7_-_150020750 | 0.12 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr12_-_91546926 | 0.12 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_-_169587621 | 0.11 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr6_-_154751629 | 0.11 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr11_-_107729504 | 0.11 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_+_133174147 | 0.11 |
ENST00000329321.3
|
GPR39
|
G protein-coupled receptor 39 |
| chr16_+_81272287 | 0.10 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr4_-_129207942 | 0.10 |
ENST00000503588.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr8_-_124749609 | 0.10 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr4_-_69434245 | 0.10 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr4_-_129208030 | 0.10 |
ENST00000503872.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr18_-_2982869 | 0.10 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr2_-_74555350 | 0.09 |
ENST00000444570.1
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr5_+_140571902 | 0.09 |
ENST00000239446.4
|
PCDHB10
|
protocadherin beta 10 |
| chr3_-_49466686 | 0.09 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr19_-_43099070 | 0.09 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr19_+_39138320 | 0.08 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr7_+_141695633 | 0.08 |
ENST00000549489.2
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr3_-_137893721 | 0.08 |
ENST00000505015.2
ENST00000260803.4 |
DBR1
|
debranching RNA lariats 1 |
| chr16_-_75284758 | 0.08 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr16_+_20775024 | 0.08 |
ENST00000289416.5
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr4_-_76944621 | 0.08 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr13_-_41593425 | 0.08 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr22_-_22337204 | 0.08 |
ENST00000430142.1
ENST00000357179.5 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr14_+_21156915 | 0.08 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr16_+_20911174 | 0.08 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr2_-_228244013 | 0.08 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr6_+_31707725 | 0.07 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr12_-_8693539 | 0.07 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr22_+_45098067 | 0.07 |
ENST00000336985.6
ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr17_+_79953310 | 0.07 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr8_-_87242589 | 0.07 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr4_-_987217 | 0.07 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr5_+_140772381 | 0.07 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr2_+_28718921 | 0.07 |
ENST00000327757.5
ENST00000422425.2 ENST00000404858.1 |
PLB1
|
phospholipase B1 |
| chr12_-_15865844 | 0.07 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr17_+_1646130 | 0.07 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr20_+_62371206 | 0.07 |
ENST00000266077.2
|
SLC2A4RG
|
SLC2A4 regulator |
| chr17_+_48823896 | 0.07 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr17_-_36904437 | 0.06 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chrX_+_120181457 | 0.06 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr22_-_22337154 | 0.06 |
ENST00000413067.2
ENST00000437929.1 ENST00000456075.1 ENST00000434517.1 ENST00000424393.1 ENST00000449704.1 ENST00000437103.1 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr9_-_94712434 | 0.06 |
ENST00000375708.3
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr7_+_141695671 | 0.06 |
ENST00000497673.1
ENST00000475668.2 |
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr19_+_29097595 | 0.06 |
ENST00000587961.1
|
AC079466.1
|
AC079466.1 |
| chr6_-_113953705 | 0.06 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr11_+_1860200 | 0.06 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr11_+_20620946 | 0.06 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr14_+_62585332 | 0.06 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr14_+_24590560 | 0.05 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr5_-_59668541 | 0.05 |
ENST00000514552.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_+_144665237 | 0.05 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr7_-_104567066 | 0.05 |
ENST00000453666.1
|
RP11-325F22.5
|
RP11-325F22.5 |
| chr9_+_22646189 | 0.05 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr3_+_183948161 | 0.05 |
ENST00000426955.2
|
VWA5B2
|
von Willebrand factor A domain containing 5B2 |
| chr19_+_54466179 | 0.05 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr5_-_111093167 | 0.05 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr5_+_67586465 | 0.05 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_+_144904334 | 0.05 |
ENST00000367526.4
|
UTRN
|
utrophin |
| chr3_+_75955817 | 0.05 |
ENST00000487694.3
ENST00000602589.1 |
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr12_+_93096759 | 0.05 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr1_+_205197304 | 0.05 |
ENST00000358024.3
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr16_-_57832004 | 0.05 |
ENST00000562503.1
|
KIFC3
|
kinesin family member C3 |
| chr1_+_207262881 | 0.05 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr17_+_73642486 | 0.05 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr22_+_32439019 | 0.04 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr19_+_39138271 | 0.04 |
ENST00000252699.2
|
ACTN4
|
actinin, alpha 4 |
| chr19_-_38916839 | 0.04 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr10_-_38146510 | 0.04 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chr10_+_111765562 | 0.04 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr7_-_143956815 | 0.04 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr13_-_103719196 | 0.04 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr12_+_60058458 | 0.04 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr6_+_126070726 | 0.04 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr4_+_108815402 | 0.04 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr1_+_17906970 | 0.04 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr9_-_123812542 | 0.04 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr17_+_41363854 | 0.04 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr21_-_47575481 | 0.03 |
ENST00000291670.5
ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD
|
formimidoyltransferase cyclodeaminase |
| chr22_-_30968839 | 0.03 |
ENST00000445645.1
ENST00000416358.1 ENST00000423371.1 ENST00000411821.1 ENST00000448604.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr1_+_104293028 | 0.03 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr11_-_117695449 | 0.03 |
ENST00000292079.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr22_-_38506619 | 0.03 |
ENST00000332536.5
ENST00000381669.3 |
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr14_-_54425475 | 0.03 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr10_-_72648541 | 0.03 |
ENST00000299299.3
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr12_+_20963632 | 0.03 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr17_+_68071389 | 0.03 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr7_+_130126012 | 0.03 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr3_+_180319918 | 0.03 |
ENST00000296015.4
ENST00000491380.1 ENST00000412756.2 ENST00000382584.4 |
TTC14
|
tetratricopeptide repeat domain 14 |
| chrX_+_1710484 | 0.03 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr6_-_161085291 | 0.03 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr4_-_70518941 | 0.03 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr22_+_31003133 | 0.03 |
ENST00000405742.3
|
TCN2
|
transcobalamin II |
| chr1_-_110284384 | 0.03 |
ENST00000540225.1
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr4_+_3443614 | 0.03 |
ENST00000382774.3
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chr2_+_234526272 | 0.03 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr10_-_38146482 | 0.02 |
ENST00000374648.3
|
ZNF248
|
zinc finger protein 248 |
| chr10_-_52645416 | 0.02 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr6_-_116833500 | 0.02 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr12_-_91574142 | 0.02 |
ENST00000547937.1
|
DCN
|
decorin |
| chr4_-_100212132 | 0.02 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chrX_-_124097620 | 0.02 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr11_-_128894053 | 0.02 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr17_+_41052808 | 0.02 |
ENST00000592383.1
ENST00000253801.2 ENST00000585489.1 |
G6PC
|
glucose-6-phosphatase, catalytic subunit |
| chr6_+_54173227 | 0.02 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr17_+_26800296 | 0.02 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr22_+_31003190 | 0.02 |
ENST00000407817.3
|
TCN2
|
transcobalamin II |
| chr19_+_41305085 | 0.02 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr1_+_66999799 | 0.02 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr7_-_47579188 | 0.02 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr7_+_130126165 | 0.02 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr9_+_140125385 | 0.02 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr3_+_107318157 | 0.02 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr19_-_11639910 | 0.02 |
ENST00000588998.1
ENST00000586149.1 |
ECSIT
|
ECSIT signalling integrator |
| chr9_+_82186872 | 0.02 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_-_36020531 | 0.02 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr7_-_27219632 | 0.02 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr11_-_83393303 | 0.02 |
ENST00000398304.1
ENST00000420775.2 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr4_-_22444733 | 0.02 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr11_-_26744908 | 0.02 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr22_-_30968813 | 0.02 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr5_+_175976324 | 0.02 |
ENST00000261944.5
|
CDHR2
|
cadherin-related family member 2 |
| chr1_+_84630645 | 0.02 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_-_54653313 | 0.02 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr8_+_20054878 | 0.02 |
ENST00000276390.2
ENST00000519667.1 |
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
| chr11_+_2405833 | 0.02 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr2_+_234627424 | 0.02 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr4_-_74486217 | 0.02 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_166034029 | 0.01 |
ENST00000306480.6
|
TMEM192
|
transmembrane protein 192 |
| chr8_-_17752996 | 0.01 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr16_-_15474904 | 0.01 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr6_-_127840336 | 0.01 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr11_+_46740730 | 0.01 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr2_-_169104651 | 0.01 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr19_+_6464243 | 0.01 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr8_+_104892639 | 0.01 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr14_-_94759361 | 0.01 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr7_-_150020578 | 0.01 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr3_-_157221128 | 0.01 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chrX_+_48660107 | 0.01 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr1_+_36789335 | 0.01 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr14_-_65409438 | 0.01 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr3_-_157221380 | 0.01 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr5_+_40841276 | 0.01 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr15_+_58430567 | 0.01 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr1_-_207206092 | 0.01 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr16_-_57831676 | 0.01 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr2_+_234621551 | 0.01 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF1A_HNF1B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.2 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.5 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.2 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.3 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 1.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.0 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |