Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
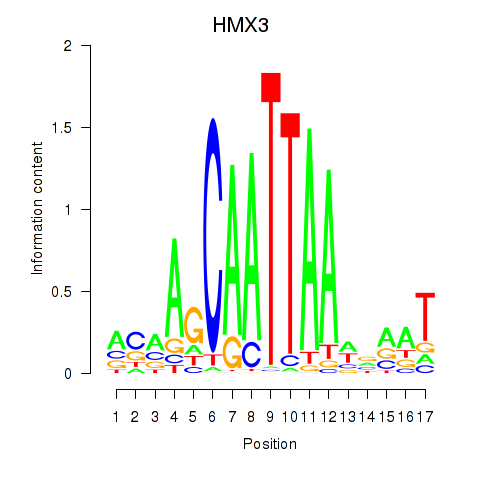
Results for HMX3
Z-value: 0.73
Transcription factors associated with HMX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX3
|
ENSG00000188620.9 | H6 family homeobox 3 |
Activity profile of HMX3 motif
Sorted Z-values of HMX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_86861660 | 0.79 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr9_-_95055923 | 0.69 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr12_+_51318513 | 0.50 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr4_+_75174180 | 0.49 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr3_+_38537960 | 0.45 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr3_-_195076933 | 0.39 |
ENST00000423531.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr5_+_146614579 | 0.36 |
ENST00000541094.1
ENST00000398521.3 |
STK32A
|
serine/threonine kinase 32A |
| chr11_+_63304273 | 0.36 |
ENST00000439013.2
ENST00000255688.3 |
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3 |
| chr19_+_9361606 | 0.35 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr7_-_35013217 | 0.34 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr5_-_142077569 | 0.33 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr7_-_77325545 | 0.32 |
ENST00000447009.1
ENST00000416650.1 ENST00000440088.1 ENST00000430801.1 ENST00000398043.2 |
RSBN1L-AS1
|
RSBN1L antisense RNA 1 |
| chrX_-_43741594 | 0.30 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr12_-_10601963 | 0.30 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr8_-_95487272 | 0.30 |
ENST00000297592.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr2_-_55237484 | 0.30 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr9_+_109685630 | 0.29 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr5_+_42756903 | 0.29 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr12_-_10022735 | 0.29 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chrX_-_55208866 | 0.28 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr8_-_95487331 | 0.28 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr9_+_123884038 | 0.28 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr4_+_83956312 | 0.28 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr16_-_56701933 | 0.28 |
ENST00000568675.1
ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G
|
metallothionein 1G |
| chr16_-_70239683 | 0.28 |
ENST00000601706.1
|
AC009060.1
|
Uncharacterized protein |
| chr15_+_42697065 | 0.27 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr5_-_42812143 | 0.27 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_+_58193388 | 0.27 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr18_+_76829441 | 0.27 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr1_+_87012922 | 0.27 |
ENST00000263723.5
|
CLCA4
|
chloride channel accessory 4 |
| chr18_+_61254534 | 0.26 |
ENST00000269489.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr1_+_160709055 | 0.26 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr16_+_57139933 | 0.26 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr2_-_68290106 | 0.26 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr20_+_5986727 | 0.25 |
ENST00000378863.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr6_-_135271219 | 0.25 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr6_-_26199471 | 0.24 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr5_-_102898465 | 0.24 |
ENST00000507423.1
ENST00000230792.2 |
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr4_-_112993808 | 0.23 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr8_-_71157595 | 0.23 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr1_-_108231101 | 0.23 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr18_+_76829385 | 0.23 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr21_+_17791648 | 0.23 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_+_130159433 | 0.23 |
ENST00000451404.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr18_+_61254570 | 0.23 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr20_+_19738300 | 0.22 |
ENST00000432334.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr7_+_39605966 | 0.22 |
ENST00000223273.2
ENST00000448268.1 ENST00000432096.2 |
YAE1D1
|
Yae1 domain containing 1 |
| chr12_+_25205568 | 0.22 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr2_+_242312264 | 0.22 |
ENST00000445489.1
|
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr12_+_57810198 | 0.22 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr14_+_96000930 | 0.22 |
ENST00000331334.4
|
GLRX5
|
glutaredoxin 5 |
| chr7_+_29603394 | 0.22 |
ENST00000319694.2
|
PRR15
|
proline rich 15 |
| chrX_+_13671225 | 0.22 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr19_+_10222189 | 0.22 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chrX_-_1331527 | 0.21 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chr2_+_97454321 | 0.21 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr4_-_39034542 | 0.21 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr5_+_147691979 | 0.21 |
ENST00000274565.4
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr16_+_70258261 | 0.21 |
ENST00000594734.1
|
FKSG63
|
FKSG63 |
| chr9_-_21482312 | 0.21 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr1_-_244006528 | 0.21 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr3_+_111697843 | 0.21 |
ENST00000534857.1
ENST00000273359.3 ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10 |
| chr14_+_50291993 | 0.20 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr5_+_95066823 | 0.20 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr5_+_112849373 | 0.20 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr22_-_29107919 | 0.19 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr14_+_23025534 | 0.19 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr2_+_90458201 | 0.19 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr16_+_53412368 | 0.19 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr9_+_71986182 | 0.19 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr15_-_60771128 | 0.19 |
ENST00000558512.1
ENST00000561114.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr1_+_87012753 | 0.19 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr19_+_507299 | 0.19 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chrX_+_119737806 | 0.19 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr21_+_25801041 | 0.19 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr11_+_65266507 | 0.19 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr19_+_42212526 | 0.18 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr19_+_40195101 | 0.18 |
ENST00000360675.3
ENST00000601802.1 |
LGALS14
|
lectin, galactoside-binding, soluble, 14 |
| chr6_-_35656685 | 0.18 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr4_+_83956237 | 0.18 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr1_+_53480598 | 0.18 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr14_+_32798547 | 0.18 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_-_152061537 | 0.18 |
ENST00000368806.1
|
TCHHL1
|
trichohyalin-like 1 |
| chr3_-_11685345 | 0.18 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr5_+_169011033 | 0.18 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr13_+_108921977 | 0.18 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr4_-_14889791 | 0.18 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr8_+_42396274 | 0.18 |
ENST00000438528.3
|
SMIM19
|
small integral membrane protein 19 |
| chr7_-_92146729 | 0.18 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr6_-_46138676 | 0.18 |
ENST00000371383.2
ENST00000230565.3 |
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr19_+_52772821 | 0.18 |
ENST00000439461.1
|
ZNF766
|
zinc finger protein 766 |
| chr4_-_104020968 | 0.18 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr8_-_27695552 | 0.17 |
ENST00000522944.1
ENST00000301905.4 |
PBK
|
PDZ binding kinase |
| chr1_-_152779104 | 0.17 |
ENST00000606576.1
ENST00000368768.1 |
LCE1C
|
late cornified envelope 1C |
| chr1_+_160709029 | 0.17 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr8_-_6115044 | 0.17 |
ENST00000519555.1
|
RP11-124B13.1
|
RP11-124B13.1 |
| chr2_+_103089756 | 0.17 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr14_+_102196739 | 0.17 |
ENST00000556973.1
|
RP11-796G6.2
|
Uncharacterized protein |
| chr19_+_41281416 | 0.17 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr6_+_26124373 | 0.17 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr4_+_25915896 | 0.17 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr1_-_110933611 | 0.17 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr1_+_160709076 | 0.17 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_171060018 | 0.17 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr10_-_61900762 | 0.17 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr6_+_134274354 | 0.17 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr11_-_62323702 | 0.17 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr19_+_45844032 | 0.17 |
ENST00000589837.1
|
KLC3
|
kinesin light chain 3 |
| chr12_+_28299014 | 0.17 |
ENST00000538586.1
ENST00000536154.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr8_-_121457332 | 0.16 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr12_+_12202785 | 0.16 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr3_+_149535022 | 0.16 |
ENST00000466795.1
|
RNF13
|
ring finger protein 13 |
| chr11_-_104893863 | 0.16 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chr1_-_149459549 | 0.16 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr1_-_160549235 | 0.16 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr12_+_128399965 | 0.16 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chrX_+_105937068 | 0.16 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr2_-_111334678 | 0.16 |
ENST00000329516.3
ENST00000330331.5 ENST00000446930.1 |
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr7_+_39773160 | 0.16 |
ENST00000340510.4
|
LINC00265
|
long intergenic non-protein coding RNA 265 |
| chr18_-_74839891 | 0.16 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr20_-_271304 | 0.15 |
ENST00000400269.3
ENST00000360321.2 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr17_-_40828969 | 0.15 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr7_+_33168856 | 0.15 |
ENST00000432983.1
|
BBS9
|
Bardet-Biedl syndrome 9 |
| chr9_-_27005686 | 0.15 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr5_+_150404904 | 0.15 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chrX_+_38420623 | 0.15 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr13_-_79233314 | 0.15 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chr15_-_56757329 | 0.15 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr18_+_2571510 | 0.15 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr4_+_159131596 | 0.15 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr5_-_74162605 | 0.15 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chrX_+_2670153 | 0.15 |
ENST00000509484.2
|
XG
|
Xg blood group |
| chr14_+_89290965 | 0.15 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr7_-_64023441 | 0.15 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr4_+_83821835 | 0.15 |
ENST00000302236.5
|
THAP9
|
THAP domain containing 9 |
| chr10_+_5238793 | 0.14 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr6_-_35656712 | 0.14 |
ENST00000357266.4
ENST00000542713.1 |
FKBP5
|
FK506 binding protein 5 |
| chr1_+_62439037 | 0.14 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr3_+_88198838 | 0.14 |
ENST00000318887.3
|
C3orf38
|
chromosome 3 open reading frame 38 |
| chr20_+_52824367 | 0.14 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr19_-_44384291 | 0.14 |
ENST00000324394.6
|
ZNF404
|
zinc finger protein 404 |
| chr4_+_109571740 | 0.14 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr10_-_99030395 | 0.14 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr14_-_74025625 | 0.14 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr3_-_170588163 | 0.14 |
ENST00000295830.8
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chrX_+_12809463 | 0.14 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr14_+_57857262 | 0.13 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr1_+_99127225 | 0.13 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr11_-_14521349 | 0.13 |
ENST00000534234.1
|
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr8_+_1993173 | 0.13 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr12_+_32115400 | 0.13 |
ENST00000381054.3
|
KIAA1551
|
KIAA1551 |
| chr12_+_7052974 | 0.13 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr4_+_56815102 | 0.13 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr20_-_45530365 | 0.13 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr12_+_128399917 | 0.13 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr17_-_40829026 | 0.13 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr6_-_26199499 | 0.13 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr3_+_121613265 | 0.12 |
ENST00000295605.2
|
SLC15A2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chrX_+_38420783 | 0.12 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr13_-_47471155 | 0.12 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr4_-_120243545 | 0.12 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_+_211342400 | 0.12 |
ENST00000417946.1
ENST00000518043.1 ENST00000523702.1 |
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr7_-_130066571 | 0.12 |
ENST00000492389.1
|
CEP41
|
centrosomal protein 41kDa |
| chr10_-_46089939 | 0.12 |
ENST00000453980.3
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr6_-_135271260 | 0.12 |
ENST00000265605.2
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr12_+_107712173 | 0.12 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr1_+_117963209 | 0.12 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr1_-_53387352 | 0.12 |
ENST00000541281.1
|
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr12_+_29302119 | 0.12 |
ENST00000536681.3
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr5_+_68665608 | 0.12 |
ENST00000509734.1
ENST00000354868.5 ENST00000521422.1 ENST00000354312.3 ENST00000345306.6 |
RAD17
|
RAD17 homolog (S. pombe) |
| chr2_-_134326009 | 0.12 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr2_-_211342292 | 0.12 |
ENST00000448951.1
|
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr1_+_186798073 | 0.12 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr19_-_22034809 | 0.12 |
ENST00000594012.1
ENST00000595461.1 ENST00000596899.1 |
ZNF43
|
zinc finger protein 43 |
| chr5_+_61601965 | 0.12 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr4_-_47465666 | 0.12 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr1_-_110933663 | 0.12 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr5_+_61602055 | 0.12 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr17_-_29641104 | 0.12 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr10_-_18948156 | 0.12 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr6_+_148663729 | 0.12 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr15_-_47426320 | 0.11 |
ENST00000557832.1
|
FKSG62
|
FKSG62 |
| chr5_+_146614679 | 0.11 |
ENST00000398523.3
|
STK32A
|
serine/threonine kinase 32A |
| chr18_-_66382289 | 0.11 |
ENST00000443099.2
ENST00000562706.1 ENST00000544714.2 |
TMX3
|
thioredoxin-related transmembrane protein 3 |
| chr12_+_122688090 | 0.11 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr8_+_134125727 | 0.11 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr7_-_112430647 | 0.11 |
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168 |
| chr1_+_170501270 | 0.11 |
ENST00000367763.3
ENST00000367762.1 |
GORAB
|
golgin, RAB6-interacting |
| chr15_+_65843130 | 0.11 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr6_-_26124138 | 0.11 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr13_-_49987885 | 0.11 |
ENST00000409082.1
|
CAB39L
|
calcium binding protein 39-like |
| chr6_+_26217159 | 0.11 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr8_-_17555164 | 0.11 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr19_-_52255107 | 0.11 |
ENST00000595042.1
ENST00000304748.4 |
FPR1
|
formyl peptide receptor 1 |
| chr9_-_5833027 | 0.11 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr1_-_54411255 | 0.11 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr14_+_65381079 | 0.11 |
ENST00000549115.1
ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1
FNTB
CHURC1-FNTB
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr11_-_64684672 | 0.11 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr7_+_91570165 | 0.11 |
ENST00000356239.3
ENST00000359028.2 ENST00000358100.2 |
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr19_+_58193337 | 0.11 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 0.5 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.1 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.3 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.0 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:0070409 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.3 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.3 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.4 | GO:1904847 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:0070269 | ectopic germ cell programmed cell death(GO:0035234) pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.7 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.6 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.0 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.0 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.0 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.0 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0098552 | side of membrane(GO:0098552) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0047756 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |