Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
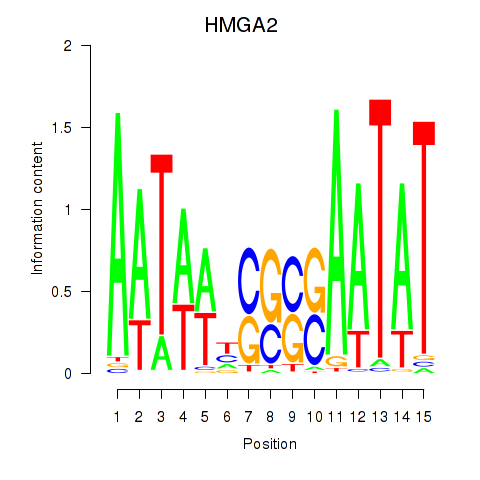
Results for HMGA2
Z-value: 0.51
Transcription factors associated with HMGA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA2
|
ENSG00000149948.9 | high mobility group AT-hook 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA2 | hg19_v2_chr12_+_66218212_66218244 | -0.90 | 1.4e-02 | Click! |
Activity profile of HMGA2 motif
Sorted Z-values of HMGA2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_65996599 | 0.53 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr14_+_50291993 | 0.21 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr1_-_67600639 | 0.19 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr3_+_121774202 | 0.18 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr6_+_116892641 | 0.18 |
ENST00000487832.2
ENST00000518117.1 |
RWDD1
|
RWD domain containing 1 |
| chr18_-_14132422 | 0.17 |
ENST00000589498.1
ENST00000590202.1 |
ZNF519
|
zinc finger protein 519 |
| chr1_+_227751231 | 0.16 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr22_+_22676808 | 0.15 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr19_+_44455368 | 0.14 |
ENST00000591168.1
ENST00000587682.1 ENST00000251269.5 |
ZNF221
|
zinc finger protein 221 |
| chr16_+_66442411 | 0.14 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr1_+_227751281 | 0.13 |
ENST00000440339.1
|
ZNF678
|
zinc finger protein 678 |
| chr12_-_10573149 | 0.12 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr6_+_111580508 | 0.12 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr13_+_50656307 | 0.11 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr11_-_18063973 | 0.10 |
ENST00000528338.1
|
TPH1
|
tryptophan hydroxylase 1 |
| chr4_+_25378912 | 0.10 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr6_-_136571400 | 0.10 |
ENST00000418509.2
ENST00000420702.1 ENST00000451457.2 |
MTFR2
|
mitochondrial fission regulator 2 |
| chr14_-_94759595 | 0.10 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr1_+_2487800 | 0.10 |
ENST00000355716.4
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr3_-_123680246 | 0.10 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr2_+_120436733 | 0.09 |
ENST00000401466.1
ENST00000424086.1 ENST00000272521.6 |
TMEM177
|
transmembrane protein 177 |
| chr2_-_287299 | 0.09 |
ENST00000405290.1
|
FAM150B
|
family with sequence similarity 150, member B |
| chr2_+_170683979 | 0.09 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr5_+_67485704 | 0.09 |
ENST00000520762.1
|
RP11-404L6.2
|
Uncharacterized protein |
| chr1_+_196621156 | 0.09 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr16_+_53483983 | 0.09 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr1_+_179263308 | 0.09 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr1_+_144989309 | 0.08 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr2_+_120436760 | 0.08 |
ENST00000445518.1
ENST00000409951.1 |
TMEM177
|
transmembrane protein 177 |
| chr10_-_62332357 | 0.08 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_196621002 | 0.08 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr20_+_54967663 | 0.08 |
ENST00000452950.1
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
| chr3_+_106959530 | 0.08 |
ENST00000466734.1
ENST00000463143.1 ENST00000490441.1 |
LINC00883
|
long intergenic non-protein coding RNA 883 |
| chr1_+_55464600 | 0.08 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr6_+_123110302 | 0.08 |
ENST00000368440.4
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr4_+_40058411 | 0.08 |
ENST00000261435.6
ENST00000515550.1 |
N4BP2
|
NEDD4 binding protein 2 |
| chr14_+_45553296 | 0.08 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr9_-_115480303 | 0.07 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr3_+_186501336 | 0.07 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr13_+_51483814 | 0.07 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr4_-_299110 | 0.07 |
ENST00000419098.1
|
ZNF732
|
zinc finger protein 732 |
| chr2_+_170683942 | 0.07 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr6_-_127664736 | 0.07 |
ENST00000368291.2
ENST00000309620.9 ENST00000454859.3 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr11_+_73661364 | 0.06 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr2_-_183387064 | 0.06 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_36608422 | 0.06 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr21_+_38792602 | 0.06 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr15_-_83018198 | 0.06 |
ENST00000557886.1
|
RP13-996F3.4
|
golgin A6 family-like 19 |
| chr12_-_122751002 | 0.06 |
ENST00000267199.4
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr2_-_287320 | 0.06 |
ENST00000401503.1
|
FAM150B
|
family with sequence similarity 150, member B |
| chr10_-_5046042 | 0.06 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chrX_-_7895755 | 0.05 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr15_-_32747835 | 0.05 |
ENST00000509311.2
ENST00000414865.2 |
GOLGA8O
|
golgin A8 family, member O |
| chr12_-_10324716 | 0.05 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr9_+_42704004 | 0.05 |
ENST00000457288.1
|
CBWD7
|
COBW domain containing 7 |
| chr15_+_84904525 | 0.05 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr18_+_11851383 | 0.05 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr15_-_70994612 | 0.05 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr6_-_127664683 | 0.05 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr15_-_82939157 | 0.05 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr1_+_64014588 | 0.04 |
ENST00000371086.2
ENST00000340052.3 |
DLEU2L
|
deleted in lymphocytic leukemia 2-like |
| chr19_-_38146289 | 0.04 |
ENST00000392144.1
ENST00000591444.1 ENST00000351218.2 ENST00000587809.1 |
ZFP30
|
ZFP30 zinc finger protein |
| chr10_+_96305535 | 0.04 |
ENST00000419900.1
ENST00000348459.5 ENST00000394045.1 ENST00000394044.1 ENST00000394036.1 |
HELLS
|
helicase, lymphoid-specific |
| chr17_-_30186328 | 0.04 |
ENST00000302362.6
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr20_+_46130671 | 0.04 |
ENST00000371998.3
ENST00000371997.3 |
NCOA3
|
nuclear receptor coactivator 3 |
| chr12_+_21207503 | 0.04 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr7_-_102985035 | 0.04 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr15_+_83098710 | 0.04 |
ENST00000561062.1
ENST00000358583.3 |
GOLGA6L9
|
golgin A6 family-like 20 |
| chr20_+_46130619 | 0.04 |
ENST00000372004.3
|
NCOA3
|
nuclear receptor coactivator 3 |
| chrX_+_2670153 | 0.04 |
ENST00000509484.2
|
XG
|
Xg blood group |
| chr4_+_184826418 | 0.04 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr1_+_2487402 | 0.04 |
ENST00000451778.1
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr9_+_104296122 | 0.04 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr11_-_130786400 | 0.04 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr10_+_90484301 | 0.03 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr18_-_29340827 | 0.03 |
ENST00000269205.5
|
SLC25A52
|
solute carrier family 25, member 52 |
| chrX_+_117480036 | 0.03 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr9_+_2157655 | 0.03 |
ENST00000452193.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_+_82722225 | 0.03 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chr14_-_94759361 | 0.03 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr2_+_177134201 | 0.03 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chrX_-_135962876 | 0.03 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chrX_+_2924654 | 0.03 |
ENST00000381130.2
|
ARSH
|
arylsulfatase family, member H |
| chr6_-_160209471 | 0.03 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr15_-_74659978 | 0.03 |
ENST00000541301.1
ENST00000416978.1 ENST00000268053.6 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr19_-_43099070 | 0.03 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr10_-_36813162 | 0.03 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr2_+_235887329 | 0.03 |
ENST00000409212.1
ENST00000344528.4 ENST00000444916.1 |
SH3BP4
|
SH3-domain binding protein 4 |
| chr1_-_76398077 | 0.03 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chrX_-_77150985 | 0.03 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr12_+_18414446 | 0.02 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr5_+_150827143 | 0.02 |
ENST00000243389.3
ENST00000517945.1 ENST00000521925.1 |
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr14_-_94759408 | 0.02 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr20_+_46130601 | 0.02 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr2_+_220144052 | 0.02 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chrX_-_7895479 | 0.02 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr6_+_31887761 | 0.02 |
ENST00000413154.1
|
C2
|
complement component 2 |
| chr7_-_102985288 | 0.01 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr15_+_88120158 | 0.01 |
ENST00000560153.1
|
LINC00052
|
long intergenic non-protein coding RNA 52 |
| chr11_-_130786333 | 0.01 |
ENST00000533214.1
ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19
|
sorting nexin 19 |
| chrX_+_2670066 | 0.01 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr2_+_172544182 | 0.01 |
ENST00000409197.1
ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr14_+_37641012 | 0.01 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr8_-_105479270 | 0.01 |
ENST00000521573.2
ENST00000351513.2 |
DPYS
|
dihydropyrimidinase |
| chr6_-_52926539 | 0.01 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr2_-_183387430 | 0.01 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_-_76220740 | 0.01 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr2_+_39117010 | 0.00 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr15_+_58702742 | 0.00 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr7_+_80275953 | 0.00 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_-_101673782 | 0.00 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.2 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |