Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
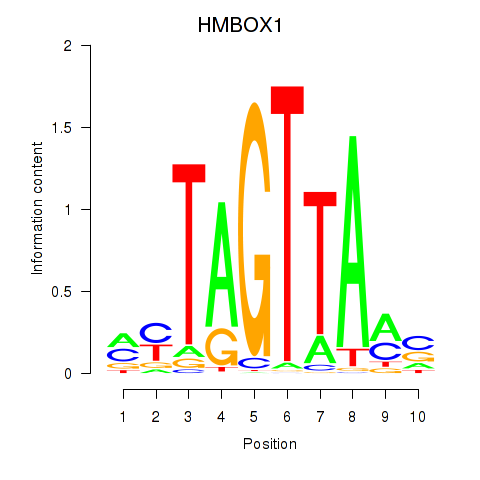
Results for HMBOX1
Z-value: 0.69
Transcription factors associated with HMBOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMBOX1
|
ENSG00000147421.13 | homeobox containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMBOX1 | hg19_v2_chr8_+_28747884_28747938 | 0.50 | 3.2e-01 | Click! |
Activity profile of HMBOX1 motif
Sorted Z-values of HMBOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_17598506 | 0.46 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr6_+_111303218 | 0.39 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr1_-_86861660 | 0.38 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr19_+_4769117 | 0.31 |
ENST00000540211.1
ENST00000317292.3 ENST00000586721.1 ENST00000592709.1 ENST00000588711.1 ENST00000589639.1 ENST00000591008.1 ENST00000592663.1 ENST00000588758.1 |
MIR7-3HG
|
MIR7-3 host gene (non-protein coding) |
| chr1_+_222988363 | 0.30 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr12_-_113658892 | 0.26 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr7_-_120498357 | 0.25 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr14_-_89878369 | 0.24 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr4_-_69434245 | 0.23 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr12_-_25348007 | 0.23 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr2_-_37501692 | 0.23 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr17_+_42925270 | 0.21 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr3_-_131756559 | 0.21 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr6_+_30585486 | 0.21 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr1_+_81001398 | 0.20 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr6_+_148593425 | 0.19 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr1_-_9970383 | 0.19 |
ENST00000400904.3
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr20_+_42187682 | 0.18 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr8_-_28347737 | 0.18 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr6_-_45544507 | 0.17 |
ENST00000563807.1
|
RP1-166H4.2
|
RP1-166H4.2 |
| chr1_+_174844645 | 0.17 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_+_107220899 | 0.17 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr18_-_53253323 | 0.17 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr4_+_25915896 | 0.17 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr8_-_67977178 | 0.17 |
ENST00000517736.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr18_-_19283649 | 0.16 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr7_+_100728720 | 0.16 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr14_-_23623577 | 0.16 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr3_-_126327398 | 0.16 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr1_+_198126209 | 0.16 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chrX_-_21776281 | 0.15 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr1_-_169396666 | 0.15 |
ENST00000456107.1
ENST00000367805.3 |
CCDC181
|
coiled-coil domain containing 181 |
| chr1_+_222988464 | 0.15 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr11_-_124981475 | 0.14 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr1_-_201346761 | 0.14 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr10_+_35484793 | 0.13 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr9_-_73029540 | 0.13 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr12_-_77272765 | 0.13 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr5_-_1799864 | 0.13 |
ENST00000510999.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr16_-_71610985 | 0.13 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr12_-_113658826 | 0.12 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr1_-_9953295 | 0.12 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr4_-_110723194 | 0.12 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr1_-_203274418 | 0.12 |
ENST00000457348.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr2_+_127413481 | 0.12 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr12_+_47473369 | 0.12 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chr4_-_114438763 | 0.12 |
ENST00000509594.1
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chrX_+_70586140 | 0.12 |
ENST00000276072.3
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr7_+_90338547 | 0.11 |
ENST00000446790.1
|
CDK14
|
cyclin-dependent kinase 14 |
| chrX_+_70586082 | 0.11 |
ENST00000373790.4
ENST00000449580.1 ENST00000423759.1 |
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr11_-_111649074 | 0.11 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr4_+_119810134 | 0.11 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr17_-_56609302 | 0.11 |
ENST00000581607.1
ENST00000317256.6 ENST00000426861.1 ENST00000580809.1 ENST00000577729.1 ENST00000583291.1 |
SEPT4
|
septin 4 |
| chr11_+_57531292 | 0.11 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr14_+_75536280 | 0.11 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr15_+_75487984 | 0.11 |
ENST00000563905.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr3_+_52444651 | 0.11 |
ENST00000327906.3
|
PHF7
|
PHD finger protein 7 |
| chr1_-_110283138 | 0.11 |
ENST00000256594.3
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr2_+_204193101 | 0.11 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chrX_+_70503526 | 0.10 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr11_-_46638720 | 0.10 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr7_+_102191679 | 0.10 |
ENST00000507918.1
ENST00000432940.1 |
SPDYE2
|
speedy/RINGO cell cycle regulator family member E2 |
| chr12_-_56236734 | 0.10 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr2_+_166430619 | 0.10 |
ENST00000409420.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr17_+_48172639 | 0.10 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr8_+_94710789 | 0.10 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr8_-_116504448 | 0.10 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_-_220110187 | 0.10 |
ENST00000295759.7
ENST00000392089.2 |
GLB1L
|
galactosidase, beta 1-like |
| chr20_+_42187608 | 0.09 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_-_152009460 | 0.09 |
ENST00000271638.2
|
S100A11
|
S100 calcium binding protein A11 |
| chr14_+_50999744 | 0.09 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr8_+_27491572 | 0.09 |
ENST00000301904.3
|
SCARA3
|
scavenger receptor class A, member 3 |
| chr11_+_100862811 | 0.09 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr8_+_70476088 | 0.09 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chrM_+_5824 | 0.09 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr1_+_207226574 | 0.09 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr19_+_38826477 | 0.09 |
ENST00000409410.2
ENST00000215069.4 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr15_+_75639372 | 0.09 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr4_+_9172135 | 0.09 |
ENST00000512047.1
|
FAM90A26
|
family with sequence similarity 90, member A26 |
| chr16_-_3422283 | 0.09 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr12_+_117013656 | 0.09 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr17_+_41177220 | 0.09 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr1_+_152748848 | 0.09 |
ENST00000334371.2
|
LCE1F
|
late cornified envelope 1F |
| chr6_+_30029008 | 0.09 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr2_-_86116020 | 0.09 |
ENST00000525834.2
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr5_+_138629628 | 0.09 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr8_-_67525473 | 0.08 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr9_+_128510454 | 0.08 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr17_-_38083092 | 0.08 |
ENST00000394169.1
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae) |
| chr22_-_37880543 | 0.08 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_-_69101413 | 0.08 |
ENST00000398559.2
|
TMF1
|
TATA element modulatory factor 1 |
| chr12_-_57113333 | 0.08 |
ENST00000550920.1
|
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr12_-_56727676 | 0.08 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr2_-_86948245 | 0.08 |
ENST00000439940.2
ENST00000604011.1 |
CHMP3
RNF103-CHMP3
|
charged multivesicular body protein 3 RNF103-CHMP3 readthrough |
| chr4_+_77941685 | 0.08 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr3_+_38347427 | 0.08 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr1_-_160549235 | 0.08 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr10_-_24911746 | 0.08 |
ENST00000320481.6
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr7_+_95115210 | 0.08 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr17_-_40134339 | 0.08 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr8_+_67976593 | 0.08 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chrX_-_69509738 | 0.08 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr10_-_112678904 | 0.08 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr6_+_96969672 | 0.08 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr2_+_61293021 | 0.08 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr2_+_192543153 | 0.08 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr1_+_116654376 | 0.08 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr7_-_102985288 | 0.08 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr7_+_128379449 | 0.08 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr6_+_112408768 | 0.07 |
ENST00000368656.2
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229, member B |
| chr4_-_110723134 | 0.07 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr2_+_204193149 | 0.07 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr6_-_52668605 | 0.07 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr1_-_21113105 | 0.07 |
ENST00000375000.1
ENST00000419490.1 ENST00000414993.1 ENST00000443615.1 ENST00000312239.5 |
HP1BP3
|
heterochromatin protein 1, binding protein 3 |
| chr3_+_159481988 | 0.07 |
ENST00000472451.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr2_+_204193129 | 0.07 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chrX_+_37208540 | 0.07 |
ENST00000466533.1
ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1
TM4SF2
|
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr1_-_150669604 | 0.07 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr1_-_149982624 | 0.07 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr4_-_159080806 | 0.07 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr13_-_67802549 | 0.07 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr14_+_75536335 | 0.07 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr15_+_75639296 | 0.07 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr7_+_102290772 | 0.07 |
ENST00000507450.1
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr8_+_120885949 | 0.06 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr18_+_11752783 | 0.06 |
ENST00000585642.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr3_-_149095652 | 0.06 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr11_+_19798964 | 0.06 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr3_-_141747439 | 0.06 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_+_65769550 | 0.06 |
ENST00000312175.2
ENST00000445560.2 ENST00000530204.1 |
BANF1
|
barrier to autointegration factor 1 |
| chr19_+_6740888 | 0.06 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr11_-_62572901 | 0.06 |
ENST00000439713.2
ENST00000531131.1 ENST00000530875.1 ENST00000531709.2 ENST00000294172.2 |
NXF1
|
nuclear RNA export factor 1 |
| chr17_+_44701402 | 0.06 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr4_+_36283213 | 0.06 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr7_-_28220354 | 0.06 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr3_-_69101461 | 0.06 |
ENST00000543976.1
|
TMF1
|
TATA element modulatory factor 1 |
| chr11_+_66115304 | 0.06 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chr19_-_7553889 | 0.06 |
ENST00000221480.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr3_-_18480173 | 0.06 |
ENST00000414509.1
|
SATB1
|
SATB homeobox 1 |
| chr19_+_38826415 | 0.06 |
ENST00000410018.1
ENST00000409235.3 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr1_+_104104379 | 0.05 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr2_-_38604398 | 0.05 |
ENST00000443098.1
ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2
|
atlastin GTPase 2 |
| chr3_+_33155525 | 0.05 |
ENST00000449224.1
|
CRTAP
|
cartilage associated protein |
| chr3_+_33155444 | 0.05 |
ENST00000320954.6
|
CRTAP
|
cartilage associated protein |
| chrX_+_70752945 | 0.05 |
ENST00000373701.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr19_-_35992780 | 0.05 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr3_+_37035289 | 0.05 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr1_+_24286287 | 0.05 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr15_+_79603479 | 0.05 |
ENST00000424155.2
ENST00000536821.1 |
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chrX_-_10851762 | 0.05 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chrX_+_70752917 | 0.05 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr4_-_90759440 | 0.05 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_-_33049690 | 0.05 |
ENST00000070846.6
ENST00000340811.4 |
PKP2
|
plakophilin 2 |
| chr19_+_57019212 | 0.05 |
ENST00000308031.5
ENST00000591537.1 |
ZNF471
|
zinc finger protein 471 |
| chr11_+_10772534 | 0.05 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr12_-_49488573 | 0.05 |
ENST00000266991.2
|
DHH
|
desert hedgehog |
| chr12_-_22063787 | 0.05 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr22_+_19419425 | 0.05 |
ENST00000333130.3
|
MRPL40
|
mitochondrial ribosomal protein L40 |
| chr5_+_131705438 | 0.05 |
ENST00000245407.3
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr5_+_176561129 | 0.05 |
ENST00000511258.1
ENST00000347982.4 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr3_-_42623805 | 0.05 |
ENST00000456515.1
|
SEC22C
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
| chr17_+_3572087 | 0.05 |
ENST00000248378.5
ENST00000397133.2 |
EMC6
|
ER membrane protein complex subunit 6 |
| chr7_+_128095945 | 0.05 |
ENST00000257696.4
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr9_-_114246635 | 0.05 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chr9_+_33290491 | 0.05 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr10_+_89419370 | 0.04 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr12_-_56727487 | 0.04 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr7_+_96634850 | 0.04 |
ENST00000518156.2
|
DLX6
|
distal-less homeobox 6 |
| chr5_+_138629417 | 0.04 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr8_-_74659693 | 0.04 |
ENST00000518767.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr8_-_101734170 | 0.04 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr14_+_78227105 | 0.04 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr2_-_62081254 | 0.04 |
ENST00000405894.3
|
FAM161A
|
family with sequence similarity 161, member A |
| chr5_-_137667459 | 0.04 |
ENST00000415130.2
ENST00000356505.3 ENST00000357274.3 ENST00000348983.3 ENST00000323760.6 |
CDC25C
|
cell division cycle 25C |
| chr7_+_101460882 | 0.04 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr2_-_3521518 | 0.04 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr7_+_132937820 | 0.04 |
ENST00000393161.2
ENST00000253861.4 |
EXOC4
|
exocyst complex component 4 |
| chr19_-_8214730 | 0.04 |
ENST00000600128.1
|
FBN3
|
fibrillin 3 |
| chr14_+_21458127 | 0.04 |
ENST00000382985.4
ENST00000556670.2 ENST00000553564.1 ENST00000554751.1 ENST00000554283.1 ENST00000555670.1 |
METTL17
|
methyltransferase like 17 |
| chr19_+_42300548 | 0.04 |
ENST00000344550.4
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr6_+_27114861 | 0.04 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr10_-_112678976 | 0.04 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr1_-_203274387 | 0.04 |
ENST00000432511.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr8_-_27630102 | 0.04 |
ENST00000356537.4
ENST00000522915.1 ENST00000539095.1 |
CCDC25
|
coiled-coil domain containing 25 |
| chr6_+_134758827 | 0.04 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr12_+_54674482 | 0.04 |
ENST00000547708.1
ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr6_+_167536230 | 0.04 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr2_-_99870744 | 0.04 |
ENST00000409238.1
ENST00000423800.1 |
LYG2
|
lysozyme G-like 2 |
| chr13_+_111365602 | 0.04 |
ENST00000333219.7
|
ING1
|
inhibitor of growth family, member 1 |
| chr5_+_176560595 | 0.04 |
ENST00000508896.1
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr2_+_36923901 | 0.04 |
ENST00000457137.2
|
VIT
|
vitrin |
| chr17_+_20483037 | 0.04 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
| chr2_-_228497888 | 0.04 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr3_+_153599269 | 0.04 |
ENST00000463297.1
|
RP11-217E22.5
|
RP11-217E22.5 |
| chr8_+_107460147 | 0.04 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr17_+_6544078 | 0.03 |
ENST00000250101.5
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr19_+_16178317 | 0.03 |
ENST00000344824.6
ENST00000538887.1 |
TPM4
|
tropomyosin 4 |
| chr7_+_128095900 | 0.03 |
ENST00000435296.2
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr6_-_130543958 | 0.03 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr8_-_95220775 | 0.03 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMBOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.4 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.0 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.0 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.0 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |