Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
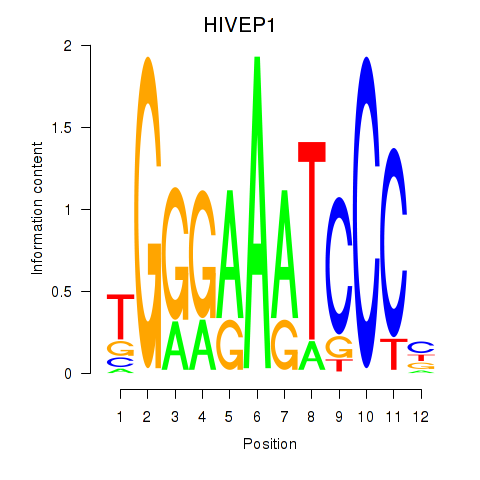
Results for HIVEP1
Z-value: 1.52
Transcription factors associated with HIVEP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIVEP1
|
ENSG00000095951.12 | HIVEP zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIVEP1 | hg19_v2_chr6_+_12012170_12012245 | -0.78 | 6.5e-02 | Click! |
Activity profile of HIVEP1 motif
Sorted Z-values of HIVEP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153066998 | 1.99 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chr4_-_74864386 | 1.87 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr1_+_37940153 | 1.59 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr5_-_150460914 | 1.50 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_+_138188351 | 1.38 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr5_+_159895275 | 1.37 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr11_+_18287801 | 1.27 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_+_18287721 | 1.26 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr14_+_103589789 | 1.22 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr17_+_40440481 | 1.21 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr6_+_138188551 | 1.05 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_-_35873856 | 1.03 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr14_+_75988851 | 0.97 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr4_+_74702214 | 0.96 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr5_-_150460539 | 0.92 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_+_138188378 | 0.83 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_+_45504688 | 0.81 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr1_+_44457441 | 0.77 |
ENST00000466180.1
|
CCDC24
|
coiled-coil domain containing 24 |
| chr3_+_177534653 | 0.75 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr17_+_77021702 | 0.74 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_6918064 | 0.70 |
ENST00000546760.1
ENST00000552402.1 |
C17orf49
|
chromosome 17 open reading frame 49 |
| chr11_-_57194111 | 0.70 |
ENST00000529112.1
ENST00000529896.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr17_+_6918093 | 0.65 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr14_-_23292596 | 0.65 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr7_+_116595028 | 0.63 |
ENST00000397751.1
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr16_+_2880157 | 0.63 |
ENST00000382280.3
|
ZG16B
|
zymogen granule protein 16B |
| chr11_-_65314905 | 0.61 |
ENST00000527339.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr17_-_39093672 | 0.59 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr15_-_41522889 | 0.58 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr6_+_33378517 | 0.57 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr11_+_64052692 | 0.56 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chr12_-_106697974 | 0.56 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr8_+_55466915 | 0.56 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr14_+_105266933 | 0.56 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr11_-_125351481 | 0.53 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr22_-_50708781 | 0.52 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr17_+_4854375 | 0.51 |
ENST00000521811.1
ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3
|
enolase 3 (beta, muscle) |
| chr14_-_24804269 | 0.50 |
ENST00000310677.4
ENST00000554068.2 ENST00000559167.1 ENST00000561138.1 |
ADCY4
|
adenylate cyclase 4 |
| chr1_+_44457261 | 0.50 |
ENST00000372318.3
|
CCDC24
|
coiled-coil domain containing 24 |
| chr10_+_124739964 | 0.50 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr17_+_6918354 | 0.50 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr17_+_40440094 | 0.49 |
ENST00000546010.2
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr4_-_76944621 | 0.49 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr19_+_45254529 | 0.49 |
ENST00000444487.1
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr17_-_62097927 | 0.47 |
ENST00000578313.1
ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2
|
intercellular adhesion molecule 2 |
| chr5_+_140579162 | 0.47 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr2_-_240230890 | 0.47 |
ENST00000446876.1
|
HDAC4
|
histone deacetylase 4 |
| chr14_-_69445793 | 0.45 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr14_-_69446034 | 0.45 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr11_-_104480019 | 0.44 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr1_+_1846519 | 0.43 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr6_+_29691198 | 0.43 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr6_+_30851205 | 0.43 |
ENST00000515881.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_+_67632083 | 0.43 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr14_-_69445968 | 0.42 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr16_+_4666475 | 0.42 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr7_-_8302298 | 0.42 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr19_-_42636543 | 0.41 |
ENST00000528894.4
ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr7_+_6713376 | 0.41 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr1_-_209824643 | 0.40 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr7_+_100271446 | 0.40 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr3_-_119396193 | 0.40 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr15_+_74509530 | 0.40 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr5_+_140552218 | 0.40 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr10_+_30723105 | 0.38 |
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr7_+_6714599 | 0.38 |
ENST00000328239.7
ENST00000542006.1 |
AC073343.1
|
Uncharacterized protein |
| chr14_+_103243813 | 0.38 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr6_+_29691056 | 0.38 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr15_-_41408409 | 0.38 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr5_+_142286887 | 0.38 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr8_+_72755367 | 0.38 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr14_+_55033815 | 0.38 |
ENST00000554335.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr1_+_114522049 | 0.37 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr9_+_97562440 | 0.37 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr7_+_100271355 | 0.37 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr14_+_55034599 | 0.36 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr17_-_18908040 | 0.35 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr12_+_52445191 | 0.35 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr19_-_17186229 | 0.35 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr19_+_4229495 | 0.34 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr14_+_20811766 | 0.34 |
ENST00000250416.5
ENST00000527915.1 |
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr1_+_41174988 | 0.34 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr17_-_6917755 | 0.34 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr17_-_53499218 | 0.34 |
ENST00000571578.1
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr19_+_17326141 | 0.34 |
ENST00000445667.2
ENST00000263897.5 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr21_+_35553045 | 0.33 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr7_-_8302164 | 0.33 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr3_-_161089289 | 0.33 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chrX_+_48367338 | 0.33 |
ENST00000359882.4
ENST00000537758.1 ENST00000367574.4 ENST00000355961.4 ENST00000489940.1 ENST00000361988.3 |
PORCN
|
porcupine homolog (Drosophila) |
| chr15_+_42066888 | 0.33 |
ENST00000510535.1
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr1_+_155051305 | 0.33 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr18_-_2982869 | 0.32 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chrX_+_10126488 | 0.32 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr3_-_100565249 | 0.32 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr22_+_50986462 | 0.31 |
ENST00000395676.2
|
KLHDC7B
|
kelch domain containing 7B |
| chr1_+_156611900 | 0.31 |
ENST00000457777.2
ENST00000424639.1 |
BCAN
|
brevican |
| chr1_+_152691998 | 0.31 |
ENST00000368775.2
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chr11_+_64052266 | 0.31 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr15_-_40398812 | 0.31 |
ENST00000561360.1
|
BMF
|
Bcl2 modifying factor |
| chr6_-_43478239 | 0.31 |
ENST00000372441.1
|
LRRC73
|
leucine rich repeat containing 73 |
| chr3_-_192445289 | 0.31 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr7_+_144015218 | 0.30 |
ENST00000408951.1
|
OR2A1
|
olfactory receptor, family 2, subfamily A, member 1 |
| chr2_-_163175133 | 0.30 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr1_+_212738676 | 0.29 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr19_+_18496957 | 0.29 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr1_-_206671061 | 0.29 |
ENST00000367119.1
|
C1orf147
|
chromosome 1 open reading frame 147 |
| chr8_-_65711310 | 0.29 |
ENST00000310193.3
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr17_+_63096903 | 0.29 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr9_-_136344197 | 0.29 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr19_-_6591113 | 0.29 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr11_+_64052944 | 0.29 |
ENST00000535675.1
ENST00000543383.1 |
GPR137
|
G protein-coupled receptor 137 |
| chr16_+_50730910 | 0.28 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr12_+_48722763 | 0.28 |
ENST00000335017.1
|
H1FNT
|
H1 histone family, member N, testis-specific |
| chr1_+_24882560 | 0.28 |
ENST00000374392.2
|
NCMAP
|
noncompact myelin associated protein |
| chr19_+_41869894 | 0.28 |
ENST00000413014.2
|
TMEM91
|
transmembrane protein 91 |
| chr11_+_64052454 | 0.28 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr19_+_35485682 | 0.28 |
ENST00000599564.1
|
GRAMD1A
|
GRAM domain containing 1A |
| chr20_+_327668 | 0.28 |
ENST00000382291.3
ENST00000609504.1 ENST00000382285.2 |
NRSN2
|
neurensin 2 |
| chr19_-_30199516 | 0.27 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr15_+_74908228 | 0.27 |
ENST00000566126.1
|
CLK3
|
CDC-like kinase 3 |
| chr14_+_20811722 | 0.27 |
ENST00000429687.3
|
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr10_+_60272814 | 0.27 |
ENST00000373886.3
|
BICC1
|
bicaudal C homolog 1 (Drosophila) |
| chr5_+_140625147 | 0.27 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr9_-_123691047 | 0.26 |
ENST00000373887.3
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr12_-_50101003 | 0.26 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr2_-_27558270 | 0.26 |
ENST00000454704.1
|
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr19_+_2270283 | 0.26 |
ENST00000588673.2
|
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr5_+_134303591 | 0.26 |
ENST00000282611.6
|
CATSPER3
|
cation channel, sperm associated 3 |
| chr19_-_37178284 | 0.26 |
ENST00000425254.2
ENST00000590952.1 ENST00000433232.1 |
AC074138.3
|
AC074138.3 |
| chr22_-_29949680 | 0.26 |
ENST00000397873.2
ENST00000490103.1 |
THOC5
|
THO complex 5 |
| chr15_+_91411810 | 0.26 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr3_-_46068969 | 0.25 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr12_-_62997214 | 0.25 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr6_-_13328050 | 0.25 |
ENST00000420456.1
|
TBC1D7
|
TBC1 domain family, member 7 |
| chr2_-_61108449 | 0.25 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr2_-_74781061 | 0.25 |
ENST00000264094.3
ENST00000393937.2 ENST00000409986.1 |
LOXL3
|
lysyl oxidase-like 3 |
| chr4_+_77177512 | 0.25 |
ENST00000606246.1
|
FAM47E
|
family with sequence similarity 47, member E |
| chr11_+_64053005 | 0.25 |
ENST00000538032.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr11_-_67276100 | 0.25 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr17_+_35306175 | 0.25 |
ENST00000225402.5
|
AATF
|
apoptosis antagonizing transcription factor |
| chr2_-_216300784 | 0.25 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr14_+_55034330 | 0.25 |
ENST00000251091.5
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr19_-_43835582 | 0.24 |
ENST00000595748.1
|
CTC-490G23.2
|
CTC-490G23.2 |
| chr6_+_139117251 | 0.24 |
ENST00000401414.3
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr6_-_31508304 | 0.24 |
ENST00000376177.2
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr5_+_53751445 | 0.24 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr5_+_127039075 | 0.24 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr5_-_149792295 | 0.24 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr12_+_57623907 | 0.24 |
ENST00000553529.1
ENST00000554310.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr6_-_33256664 | 0.24 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr1_+_15272271 | 0.24 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr17_-_9479128 | 0.24 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr19_+_51897742 | 0.23 |
ENST00000600765.1
|
CTD-2616J11.14
|
CTD-2616J11.14 |
| chr19_-_51466681 | 0.23 |
ENST00000456750.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr15_+_57891609 | 0.23 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr3_+_9774164 | 0.23 |
ENST00000426583.1
|
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr2_-_113999260 | 0.23 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chr19_+_41256764 | 0.23 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr15_+_59730348 | 0.23 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr1_+_110453203 | 0.23 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr6_-_33290580 | 0.22 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr6_-_31324943 | 0.22 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr1_-_21616690 | 0.22 |
ENST00000264205.6
|
ECE1
|
endothelin converting enzyme 1 |
| chr12_+_10365082 | 0.22 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr2_-_220252530 | 0.22 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr12_+_112451222 | 0.22 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr15_+_74908147 | 0.22 |
ENST00000568139.1
ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3
|
CDC-like kinase 3 |
| chr6_-_112115074 | 0.22 |
ENST00000368667.2
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr15_+_91427726 | 0.22 |
ENST00000452243.1
|
FES
|
feline sarcoma oncogene |
| chr6_-_6711235 | 0.22 |
ENST00000432823.2
|
RP1-80N2.2
|
RP1-80N2.2 |
| chr3_+_187086120 | 0.21 |
ENST00000259030.2
|
RTP4
|
receptor (chemosensory) transporter protein 4 |
| chr22_-_29949634 | 0.21 |
ENST00000397872.1
ENST00000397871.1 ENST00000440771.1 |
THOC5
|
THO complex 5 |
| chr16_-_85969774 | 0.21 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr7_-_22259845 | 0.21 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr9_-_112970436 | 0.21 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr18_-_72920372 | 0.21 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr7_+_18548878 | 0.20 |
ENST00000456174.2
|
HDAC9
|
histone deacetylase 9 |
| chr16_+_88772866 | 0.20 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr17_-_1389228 | 0.20 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr2_+_74120094 | 0.20 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr21_+_34775698 | 0.20 |
ENST00000381995.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr6_-_32908792 | 0.20 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chrX_+_47444613 | 0.20 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr7_-_87849340 | 0.20 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr17_+_7461580 | 0.20 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr8_+_27168988 | 0.20 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr17_-_53499310 | 0.19 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr19_-_51522955 | 0.19 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr19_-_5978090 | 0.19 |
ENST00000592621.1
ENST00000034275.8 ENST00000591092.1 ENST00000591333.1 ENST00000590623.1 ENST00000439268.2 ENST00000587159.1 |
RANBP3
|
RAN binding protein 3 |
| chr16_+_75032901 | 0.19 |
ENST00000335325.4
ENST00000320619.6 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr13_-_46756351 | 0.19 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr14_-_24664540 | 0.19 |
ENST00000530563.1
ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chrX_+_9935392 | 0.19 |
ENST00000445307.2
|
AC002365.1
|
Homo sapiens uncharacterized LOC100288814 (LOC100288814), mRNA. |
| chr19_-_44324750 | 0.19 |
ENST00000594049.1
ENST00000414615.2 |
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr12_+_57624059 | 0.18 |
ENST00000557427.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr14_-_69262916 | 0.18 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr4_-_122085469 | 0.18 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr19_-_4831701 | 0.18 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr6_-_44281043 | 0.18 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr5_-_58652788 | 0.18 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of HIVEP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.5 | 1.6 | GO:2000625 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.5 | 2.4 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.3 | 1.7 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 1.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.2 | 0.5 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.2 | 0.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 3.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.6 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.3 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.5 | GO:1903974 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.6 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.9 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.5 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 2.9 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 0.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.3 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.4 | GO:0038155 | positive regulation of activation of Janus kinase activity(GO:0010536) interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 0.2 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 | 0.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.2 | GO:0072304 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.2 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.9 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.7 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0061386 | noradrenergic neuron differentiation(GO:0003357) closure of optic fissure(GO:0061386) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 1.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.1 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.8 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.4 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 2.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.3 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0035137 | embryonic hindlimb morphogenesis(GO:0035116) hindlimb morphogenesis(GO:0035137) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:1904339 | superior temporal gyrus development(GO:0071109) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.5 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.2 | GO:0048298 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0051709 | regulation of killing of cells of other organism(GO:0051709) positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.0 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.2 | GO:0009799 | determination of left/right symmetry(GO:0007368) specification of symmetry(GO:0009799) determination of bilateral symmetry(GO:0009855) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.6 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.4 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 2.6 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 1.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 3.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 3.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 0.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.8 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.9 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.4 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 1.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.1 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 2.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 1.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 1.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) porin activity(GO:0015288) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 7.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 4.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |