Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
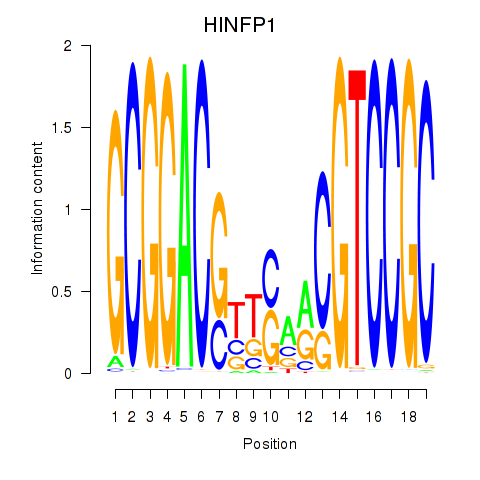
Results for HINFP1
Z-value: 0.25
Transcription factors associated with HINFP1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of HINFP1 motif
Sorted Z-values of HINFP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_92219337 | 0.14 |
ENST00000456502.1
ENST00000427372.1 |
FAM133B
|
family with sequence similarity 133, member B |
| chr1_+_42922173 | 0.12 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr2_+_130939827 | 0.12 |
ENST00000409255.1
ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr4_+_2043689 | 0.12 |
ENST00000382878.3
ENST00000409248.4 |
C4orf48
|
chromosome 4 open reading frame 48 |
| chr1_+_42921761 | 0.11 |
ENST00000372562.1
|
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr18_+_11981547 | 0.11 |
ENST00000588927.1
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr19_+_33685490 | 0.10 |
ENST00000253193.7
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr10_+_22614547 | 0.10 |
ENST00000416820.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr6_-_56707943 | 0.09 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr1_+_109102652 | 0.08 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr9_-_94186131 | 0.07 |
ENST00000297689.3
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr2_+_130939235 | 0.07 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr6_-_168720382 | 0.06 |
ENST00000610183.1
ENST00000607983.1 ENST00000366795.3 |
DACT2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr14_+_73704201 | 0.06 |
ENST00000340738.5
ENST00000427855.1 ENST00000381166.3 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr15_-_32162833 | 0.06 |
ENST00000560598.1
|
OTUD7A
|
OTU domain containing 7A |
| chr18_+_11981427 | 0.06 |
ENST00000269159.3
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr2_-_234763147 | 0.05 |
ENST00000411486.2
ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP
|
Holliday junction recognition protein |
| chr13_-_22033392 | 0.05 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr3_+_133292759 | 0.05 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr4_-_2043630 | 0.04 |
ENST00000455762.1
|
NELFA
|
negative elongation factor complex member A |
| chr3_-_196045127 | 0.04 |
ENST00000325318.5
|
TCTEX1D2
|
Tctex1 domain containing 2 |
| chr1_-_222886526 | 0.04 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr4_+_2043777 | 0.03 |
ENST00000409860.1
|
C4orf48
|
chromosome 4 open reading frame 48 |
| chr10_+_49514698 | 0.03 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr10_+_72238517 | 0.03 |
ENST00000263563.6
|
PALD1
|
phosphatase domain containing, paladin 1 |
| chr9_-_33264676 | 0.03 |
ENST00000472232.3
ENST00000379704.2 |
BAG1
|
BCL2-associated athanogene |
| chr22_+_27053422 | 0.03 |
ENST00000413665.1
ENST00000421151.1 ENST00000456129.1 ENST00000430080.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr2_-_234763105 | 0.02 |
ENST00000454020.1
|
HJURP
|
Holliday junction recognition protein |
| chr1_+_222886694 | 0.02 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr19_-_5680499 | 0.02 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr14_+_45366518 | 0.02 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr19_+_1524072 | 0.02 |
ENST00000454744.2
|
PLK5
|
polo-like kinase 5 |
| chr16_+_57279248 | 0.01 |
ENST00000562023.1
ENST00000563234.1 |
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr19_+_4198072 | 0.01 |
ENST00000262970.5
|
ANKRD24
|
ankyrin repeat domain 24 |
| chr7_+_1609694 | 0.01 |
ENST00000437621.2
ENST00000457484.2 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr3_+_133293278 | 0.00 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr1_-_62785054 | 0.00 |
ENST00000371153.4
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr1_+_210111534 | 0.00 |
ENST00000422431.1
ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14
|
synaptotagmin XIV |
| chr11_+_57435441 | 0.00 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |