Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
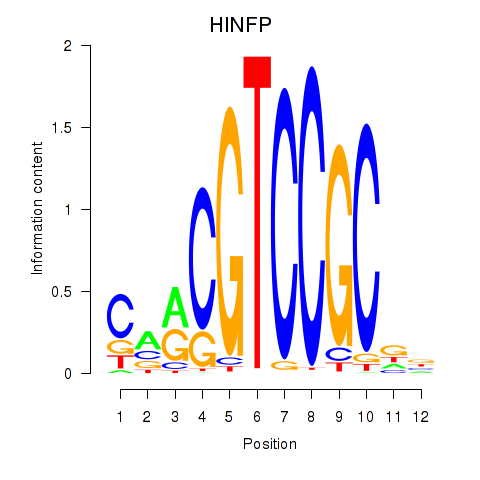
Results for HINFP
Z-value: 0.82
Transcription factors associated with HINFP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HINFP
|
ENSG00000172273.8 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HINFP | hg19_v2_chr11_+_118992269_118992334 | -0.72 | 1.1e-01 | Click! |
Activity profile of HINFP motif
Sorted Z-values of HINFP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_54963026 | 0.56 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr16_-_54962625 | 0.55 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr4_-_186732892 | 0.29 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_54962415 | 0.27 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr1_-_92952433 | 0.26 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr4_+_166128735 | 0.23 |
ENST00000226725.6
|
KLHL2
|
kelch-like family member 2 |
| chr8_+_32406179 | 0.21 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr1_-_16563641 | 0.20 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr8_+_32405728 | 0.20 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr8_+_32405785 | 0.20 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr12_+_12510352 | 0.19 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr14_-_81902791 | 0.19 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr14_-_81902516 | 0.19 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr17_-_73178599 | 0.18 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr17_-_1419941 | 0.18 |
ENST00000498390.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr22_+_29168652 | 0.18 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr3_-_57583185 | 0.18 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr4_-_183838372 | 0.17 |
ENST00000503820.1
ENST00000503988.1 |
DCTD
|
dCMP deaminase |
| chr22_+_32871224 | 0.17 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr14_+_75894714 | 0.17 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr9_-_131085021 | 0.17 |
ENST00000372890.4
|
TRUB2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr10_-_62761188 | 0.16 |
ENST00000357917.4
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr6_+_24495067 | 0.16 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr3_-_196295437 | 0.16 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr12_+_70637494 | 0.16 |
ENST00000548159.1
ENST00000549750.1 ENST00000551043.1 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr10_-_75532373 | 0.15 |
ENST00000595757.1
|
AC022400.2
|
Uncharacterized protein; cDNA FLJ44715 fis, clone BRACE3021430 |
| chr13_-_31040060 | 0.15 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr4_-_183838596 | 0.15 |
ENST00000508994.1
ENST00000512766.1 |
DCTD
|
dCMP deaminase |
| chr22_+_32870661 | 0.15 |
ENST00000266087.7
|
FBXO7
|
F-box protein 7 |
| chr20_+_30458431 | 0.14 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr1_+_109102652 | 0.14 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr2_+_30670209 | 0.14 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr3_-_150481218 | 0.14 |
ENST00000482706.1
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr3_-_184870751 | 0.13 |
ENST00000335012.2
|
C3orf70
|
chromosome 3 open reading frame 70 |
| chr13_+_21714653 | 0.13 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr5_+_36606992 | 0.13 |
ENST00000505202.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chrX_+_154299690 | 0.13 |
ENST00000340647.4
ENST00000330045.7 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr17_-_42276574 | 0.13 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr11_-_75062829 | 0.13 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr6_+_26104104 | 0.12 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr18_-_44336998 | 0.12 |
ENST00000315087.7
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr4_-_183838747 | 0.12 |
ENST00000438320.2
|
DCTD
|
dCMP deaminase |
| chr7_+_97910962 | 0.12 |
ENST00000539286.1
|
BRI3
|
brain protein I3 |
| chr17_-_1419914 | 0.12 |
ENST00000397335.3
ENST00000574561.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr8_+_32406137 | 0.11 |
ENST00000521670.1
|
NRG1
|
neuregulin 1 |
| chr1_-_1509931 | 0.11 |
ENST00000359060.4
|
SSU72
|
SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) |
| chr9_+_139921916 | 0.11 |
ENST00000314330.2
|
C9orf139
|
chromosome 9 open reading frame 139 |
| chr6_+_24495185 | 0.11 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr2_+_42396472 | 0.11 |
ENST00000318522.5
ENST00000402711.2 |
EML4
|
echinoderm microtubule associated protein like 4 |
| chr19_-_14016877 | 0.11 |
ENST00000454313.1
ENST00000591586.1 ENST00000346736.2 |
C19orf57
|
chromosome 19 open reading frame 57 |
| chr12_+_70637192 | 0.11 |
ENST00000550160.1
ENST00000551132.1 ENST00000552915.1 ENST00000552483.1 ENST00000550641.1 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr12_+_12764773 | 0.11 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr4_-_183838482 | 0.11 |
ENST00000357067.3
ENST00000510370.1 |
DCTD
|
dCMP deaminase |
| chr3_-_52312337 | 0.11 |
ENST00000469000.1
|
WDR82
|
WD repeat domain 82 |
| chr8_-_102218292 | 0.11 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr15_-_83953466 | 0.10 |
ENST00000345382.2
|
BNC1
|
basonuclin 1 |
| chr2_+_42396574 | 0.10 |
ENST00000401738.3
|
EML4
|
echinoderm microtubule associated protein like 4 |
| chr5_+_65440032 | 0.10 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr11_-_75062730 | 0.10 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr11_-_46867780 | 0.10 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr5_-_127873496 | 0.10 |
ENST00000508989.1
|
FBN2
|
fibrillin 2 |
| chr19_+_45596398 | 0.10 |
ENST00000544069.2
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr3_-_52719810 | 0.10 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr22_+_23487513 | 0.10 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr17_-_73179046 | 0.10 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chrX_+_154299753 | 0.09 |
ENST00000369459.2
ENST00000369462.1 ENST00000411985.1 ENST00000399042.1 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr13_+_21714711 | 0.09 |
ENST00000607003.1
ENST00000492245.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr17_-_42277203 | 0.09 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr5_+_142149932 | 0.09 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr8_-_100905363 | 0.09 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr3_-_196295385 | 0.09 |
ENST00000425888.1
|
WDR53
|
WD repeat domain 53 |
| chr1_+_212208919 | 0.09 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr17_+_30771279 | 0.09 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr17_-_48450534 | 0.09 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr1_+_29063119 | 0.09 |
ENST00000474884.1
ENST00000542507.1 |
YTHDF2
|
YTH domain family, member 2 |
| chr5_+_142149955 | 0.09 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr12_+_56522001 | 0.09 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr12_+_90102729 | 0.09 |
ENST00000605386.1
|
LINC00936
|
long intergenic non-protein coding RNA 936 |
| chr4_-_186733363 | 0.09 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_144329531 | 0.09 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr19_-_35323762 | 0.09 |
ENST00000590963.1
|
CTC-523E23.4
|
CTC-523E23.4 |
| chr17_+_11501816 | 0.09 |
ENST00000454412.2
|
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr9_+_37120536 | 0.09 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr3_+_32433154 | 0.09 |
ENST00000334983.5
ENST00000349718.4 |
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr3_+_110790867 | 0.09 |
ENST00000486596.1
ENST00000493615.1 |
PVRL3
|
poliovirus receptor-related 3 |
| chr4_+_148402069 | 0.08 |
ENST00000358556.4
ENST00000339690.5 ENST00000511804.1 ENST00000324300.5 |
EDNRA
|
endothelin receptor type A |
| chr17_-_4269768 | 0.08 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr10_+_99079008 | 0.08 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr4_-_107237374 | 0.08 |
ENST00000361687.4
ENST00000507696.1 ENST00000394708.2 ENST00000509532.1 |
TBCK
|
TBC1 domain containing kinase |
| chr1_+_32687971 | 0.08 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr7_-_140624499 | 0.08 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr17_-_16395455 | 0.08 |
ENST00000409083.3
|
FAM211A
|
family with sequence similarity 211, member A |
| chr3_+_133524459 | 0.08 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr2_+_157292859 | 0.08 |
ENST00000438166.2
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr1_-_40157345 | 0.08 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr14_-_102026643 | 0.08 |
ENST00000555882.1
ENST00000554441.1 ENST00000553729.1 ENST00000557109.1 ENST00000557532.1 ENST00000554694.1 ENST00000554735.1 ENST00000555174.1 ENST00000557661.1 |
DIO3OS
|
DIO3 opposite strand/antisense RNA (head to head) |
| chr10_+_75532028 | 0.08 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr16_+_70557685 | 0.08 |
ENST00000302516.5
ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3
|
splicing factor 3b, subunit 3, 130kDa |
| chr3_-_49377446 | 0.08 |
ENST00000351842.4
ENST00000416417.1 ENST00000415188.1 |
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr12_+_12509990 | 0.07 |
ENST00000542728.1
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr3_-_49449350 | 0.07 |
ENST00000454011.2
ENST00000445425.1 ENST00000422781.1 |
RHOA
|
ras homolog family member A |
| chr16_-_2162831 | 0.07 |
ENST00000483024.1
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr11_-_6633799 | 0.07 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr3_+_183853052 | 0.07 |
ENST00000273783.3
ENST00000432569.1 ENST00000444495.1 |
EIF2B5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa |
| chr16_-_70557430 | 0.07 |
ENST00000393612.4
ENST00000564653.1 ENST00000323786.5 |
COG4
|
component of oligomeric golgi complex 4 |
| chr6_+_56819895 | 0.07 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr12_+_53773944 | 0.07 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr7_-_117513540 | 0.07 |
ENST00000160373.3
|
CTTNBP2
|
cortactin binding protein 2 |
| chr10_-_118502070 | 0.07 |
ENST00000369209.3
|
HSPA12A
|
heat shock 70kDa protein 12A |
| chr13_+_25875785 | 0.07 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr4_+_154387480 | 0.07 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr6_-_144329384 | 0.07 |
ENST00000417959.2
|
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr14_-_23504087 | 0.07 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr2_+_30670127 | 0.07 |
ENST00000540623.1
ENST00000476038.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr6_+_159291090 | 0.07 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chrX_-_134232630 | 0.07 |
ENST00000535837.1
ENST00000433425.2 |
LINC00087
|
long intergenic non-protein coding RNA 87 |
| chr2_-_200335983 | 0.07 |
ENST00000457245.1
|
SATB2
|
SATB homeobox 2 |
| chr1_-_36916066 | 0.07 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr9_-_74383302 | 0.06 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr10_+_94608218 | 0.06 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr19_+_917287 | 0.06 |
ENST00000592648.1
ENST00000234371.5 |
KISS1R
|
KISS1 receptor |
| chr2_+_99953816 | 0.06 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chrX_-_15353629 | 0.06 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr3_-_13009168 | 0.06 |
ENST00000273221.4
|
IQSEC1
|
IQ motif and Sec7 domain 1 |
| chr11_-_2162162 | 0.06 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_110790715 | 0.06 |
ENST00000319792.3
|
PVRL3
|
poliovirus receptor-related 3 |
| chr3_-_129612394 | 0.06 |
ENST00000505616.1
ENST00000426664.2 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr16_+_14726672 | 0.06 |
ENST00000261658.2
ENST00000563971.1 |
BFAR
|
bifunctional apoptosis regulator |
| chr22_+_18593097 | 0.06 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr7_+_94023873 | 0.06 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr4_+_89514516 | 0.06 |
ENST00000452979.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr7_-_29234802 | 0.06 |
ENST00000449801.1
ENST00000409850.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr3_-_49449521 | 0.06 |
ENST00000431929.1
ENST00000418115.1 |
RHOA
|
ras homolog family member A |
| chr19_+_6372444 | 0.06 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr20_-_39946237 | 0.06 |
ENST00000441102.2
ENST00000559234.1 |
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr1_-_1510204 | 0.06 |
ENST00000291386.3
|
SSU72
|
SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) |
| chr14_+_52118694 | 0.06 |
ENST00000554778.1
|
FRMD6
|
FERM domain containing 6 |
| chr17_-_66453562 | 0.06 |
ENST00000262139.5
ENST00000546360.1 |
WIPI1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr2_+_121010370 | 0.06 |
ENST00000420510.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr3_-_52312636 | 0.06 |
ENST00000296490.3
|
WDR82
|
WD repeat domain 82 |
| chr22_-_22221658 | 0.06 |
ENST00000544786.1
|
MAPK1
|
mitogen-activated protein kinase 1 |
| chr6_+_56820018 | 0.06 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr1_+_214454492 | 0.06 |
ENST00000366957.5
ENST00000415093.2 |
SMYD2
|
SET and MYND domain containing 2 |
| chr2_-_97304105 | 0.06 |
ENST00000599854.1
ENST00000441706.2 |
KANSL3
|
KAT8 regulatory NSL complex subunit 3 |
| chr3_+_63897605 | 0.06 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr9_-_123555655 | 0.06 |
ENST00000340778.5
ENST00000453291.1 ENST00000608872.1 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr11_-_2162468 | 0.06 |
ENST00000434045.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr4_-_74124502 | 0.06 |
ENST00000358602.4
ENST00000330838.6 ENST00000561029.1 |
ANKRD17
|
ankyrin repeat domain 17 |
| chr1_+_29063439 | 0.06 |
ENST00000541996.1
ENST00000496288.1 |
YTHDF2
|
YTH domain family, member 2 |
| chr3_-_49377499 | 0.06 |
ENST00000265560.4
|
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr17_-_48450265 | 0.06 |
ENST00000507088.1
|
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr13_-_31039375 | 0.05 |
ENST00000399494.1
|
HMGB1
|
high mobility group box 1 |
| chr1_-_92351666 | 0.05 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chr1_+_28995231 | 0.05 |
ENST00000373816.1
|
GMEB1
|
glucocorticoid modulatory element binding protein 1 |
| chr7_+_156931606 | 0.05 |
ENST00000348165.5
|
UBE3C
|
ubiquitin protein ligase E3C |
| chr5_-_154317740 | 0.05 |
ENST00000285873.7
|
GEMIN5
|
gem (nuclear organelle) associated protein 5 |
| chr9_-_19380196 | 0.05 |
ENST00000315377.4
ENST00000380384.1 ENST00000380381.3 ENST00000380394.4 |
RPS6
|
ribosomal protein S6 |
| chr4_+_89514402 | 0.05 |
ENST00000426683.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr3_-_38691119 | 0.05 |
ENST00000333535.4
ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit |
| chrX_-_154299501 | 0.05 |
ENST00000369476.3
ENST00000369484.3 |
MTCP1
CMC4
|
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chr14_-_99947168 | 0.05 |
ENST00000331768.5
|
SETD3
|
SET domain containing 3 |
| chr8_+_110656344 | 0.05 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
| chr7_-_76256557 | 0.05 |
ENST00000275569.4
ENST00000310842.4 |
POMZP3
|
POM121 and ZP3 fusion |
| chr18_+_42260059 | 0.05 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr12_+_65672702 | 0.05 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr14_-_24768913 | 0.05 |
ENST00000288111.7
|
DHRS1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr7_+_97910981 | 0.05 |
ENST00000297290.3
|
BRI3
|
brain protein I3 |
| chr18_+_43914159 | 0.05 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr13_+_21714913 | 0.05 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr12_+_51985001 | 0.05 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr16_-_28074822 | 0.05 |
ENST00000395724.3
ENST00000380898.2 ENST00000447459.2 |
GSG1L
|
GSG1-like |
| chr8_-_102217515 | 0.05 |
ENST00000520347.1
ENST00000523922.1 ENST00000520984.1 |
ZNF706
|
zinc finger protein 706 |
| chr9_+_77112244 | 0.05 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr3_+_32433363 | 0.05 |
ENST00000465248.1
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr5_-_41510656 | 0.05 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr15_+_89346699 | 0.05 |
ENST00000558207.1
|
ACAN
|
aggrecan |
| chr11_-_62446527 | 0.05 |
ENST00000294119.2
ENST00000529640.1 ENST00000534176.1 ENST00000301935.5 |
UBXN1
|
UBX domain protein 1 |
| chr5_-_132073210 | 0.05 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr5_+_137688285 | 0.05 |
ENST00000314358.5
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr12_+_72666407 | 0.05 |
ENST00000261180.4
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme |
| chr4_-_186733119 | 0.05 |
ENST00000419063.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_+_4526341 | 0.05 |
ENST00000458134.3
ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2
|
heme oxygenase (decycling) 2 |
| chr14_-_101034811 | 0.05 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr17_-_46115122 | 0.05 |
ENST00000006101.4
|
COPZ2
|
coatomer protein complex, subunit zeta 2 |
| chr11_+_119076745 | 0.04 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr8_-_100905925 | 0.04 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr3_+_152552685 | 0.04 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr17_-_4544960 | 0.04 |
ENST00000293761.3
|
ALOX15
|
arachidonate 15-lipoxygenase |
| chr6_+_44355257 | 0.04 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr11_+_47586982 | 0.04 |
ENST00000426530.2
ENST00000534775.1 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr4_+_107236847 | 0.04 |
ENST00000358008.3
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr5_-_132166579 | 0.04 |
ENST00000378679.3
|
SHROOM1
|
shroom family member 1 |
| chr22_-_28197486 | 0.04 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr17_+_12693001 | 0.04 |
ENST00000262444.9
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr6_+_56819773 | 0.04 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr5_-_132166303 | 0.04 |
ENST00000440118.1
|
SHROOM1
|
shroom family member 1 |
| chrX_+_40944871 | 0.04 |
ENST00000378308.2
ENST00000324545.8 |
USP9X
|
ubiquitin specific peptidase 9, X-linked |
| chr8_-_97173020 | 0.04 |
ENST00000287020.5
|
GDF6
|
growth differentiation factor 6 |
| chr14_+_52118576 | 0.04 |
ENST00000395718.2
ENST00000344768.5 |
FRMD6
|
FERM domain containing 6 |
| chr17_+_48450575 | 0.04 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.7 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.6 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.1 | 0.3 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.1 | 0.2 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.0 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.2 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.3 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0070777 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.0 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) regulation of penile erection(GO:0060405) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.1 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |