Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
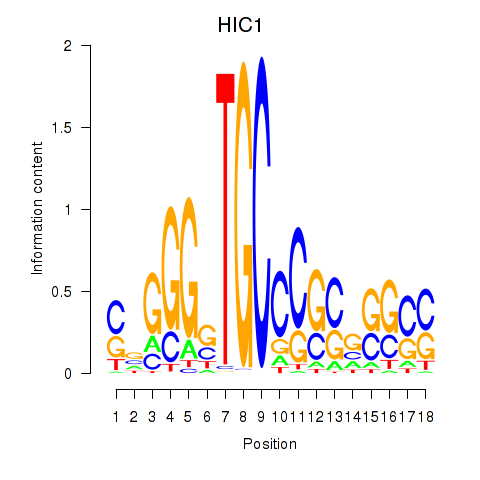
Results for HIC1
Z-value: 0.75
Transcription factors associated with HIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIC1
|
ENSG00000177374.8 | HIC ZBTB transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIC1 | hg19_v2_chr17_+_1959369_1959604 | -0.44 | 3.8e-01 | Click! |
Activity profile of HIC1 motif
Sorted Z-values of HIC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_23951673 | 0.59 |
ENST00000507936.1
|
C5orf17
|
chromosome 5 open reading frame 17 |
| chr19_-_18717627 | 0.57 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr2_-_132249955 | 0.39 |
ENST00000309451.6
|
MZT2A
|
mitotic spindle organizing protein 2A |
| chr11_-_62389577 | 0.35 |
ENST00000534715.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr11_+_65779283 | 0.31 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr20_+_49411543 | 0.30 |
ENST00000609336.1
ENST00000445038.1 |
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr16_+_619931 | 0.30 |
ENST00000321878.5
ENST00000439574.1 ENST00000026218.5 ENST00000470411.2 |
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr19_+_708910 | 0.28 |
ENST00000264560.7
|
PALM
|
paralemmin |
| chr5_+_176873789 | 0.28 |
ENST00000323249.3
ENST00000502922.1 |
PRR7
|
proline rich 7 (synaptic) |
| chr19_-_663277 | 0.27 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chr2_+_130939827 | 0.26 |
ENST00000409255.1
ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr10_-_126432619 | 0.26 |
ENST00000337318.3
|
FAM53B
|
family with sequence similarity 53, member B |
| chr9_-_138987115 | 0.26 |
ENST00000277554.2
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing |
| chr19_+_33685490 | 0.26 |
ENST00000253193.7
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr13_-_103426081 | 0.25 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr8_+_145726472 | 0.25 |
ENST00000528430.1
|
PPP1R16A
|
protein phosphatase 1, regulatory subunit 16A |
| chrX_-_45710920 | 0.25 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr4_+_183370146 | 0.24 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr19_+_54372877 | 0.24 |
ENST00000414489.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr1_-_48462566 | 0.24 |
ENST00000606738.2
|
TRABD2B
|
TraB domain containing 2B |
| chr2_+_74212073 | 0.22 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr3_+_49027771 | 0.22 |
ENST00000475629.1
ENST00000444213.1 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr19_+_18530146 | 0.22 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr14_+_65170820 | 0.22 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_+_52279737 | 0.22 |
ENST00000457351.2
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr1_+_22889953 | 0.22 |
ENST00000374644.4
ENST00000166244.3 ENST00000538803.1 |
EPHA8
|
EPH receptor A8 |
| chr4_-_2420335 | 0.22 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr12_-_108154705 | 0.22 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr20_-_62462566 | 0.21 |
ENST00000245663.4
ENST00000302995.2 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr12_+_132312931 | 0.21 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr15_-_90645679 | 0.21 |
ENST00000539790.1
ENST00000559482.1 ENST00000330062.3 |
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr12_+_47473369 | 0.21 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chrX_-_102565858 | 0.21 |
ENST00000449185.1
ENST00000536889.1 |
BEX2
|
brain expressed X-linked 2 |
| chr12_-_42631529 | 0.20 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr19_-_2236290 | 0.20 |
ENST00000591099.2
ENST00000586608.2 ENST00000326631.2 ENST00000587962.2 |
PLEKHJ1
|
pleckstrin homology domain containing, family J member 1 |
| chr1_+_1551220 | 0.20 |
ENST00000378712.1
|
MIB2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr4_-_15683230 | 0.20 |
ENST00000515679.1
|
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr10_+_71562180 | 0.19 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr3_-_49395892 | 0.19 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chr1_-_119543994 | 0.19 |
ENST00000439394.1
ENST00000449439.1 |
RP4-712E4.1
RP4-712E4.2
|
RP4-712E4.1 RP4-712E4.2 |
| chrX_-_102565932 | 0.19 |
ENST00000372674.1
ENST00000372677.3 |
BEX2
|
brain expressed X-linked 2 |
| chr17_+_74261413 | 0.18 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr16_-_838329 | 0.18 |
ENST00000563560.1
ENST00000569601.1 ENST00000565809.1 ENST00000565377.1 ENST00000007264.2 ENST00000567114.1 |
RPUSD1
|
RNA pseudouridylate synthase domain containing 1 |
| chr16_+_88519669 | 0.18 |
ENST00000319555.3
|
ZFPM1
|
zinc finger protein, FOG family member 1 |
| chr5_+_76506706 | 0.18 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr9_-_101471479 | 0.18 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr4_+_2043689 | 0.18 |
ENST00000382878.3
ENST00000409248.4 |
C4orf48
|
chromosome 4 open reading frame 48 |
| chr19_-_18391708 | 0.18 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr17_+_73084038 | 0.18 |
ENST00000578376.1
ENST00000329783.4 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr10_-_46168156 | 0.17 |
ENST00000374371.2
ENST00000335258.7 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr13_-_103426112 | 0.17 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr22_+_46476192 | 0.17 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr12_-_53574671 | 0.17 |
ENST00000444623.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr17_+_27894180 | 0.17 |
ENST00000583940.1
|
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr16_+_67563250 | 0.17 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr8_+_99956662 | 0.17 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr19_-_460996 | 0.17 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr7_+_2559399 | 0.17 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr18_+_70536215 | 0.17 |
ENST00000578967.1
|
RP11-676J15.1
|
RP11-676J15.1 |
| chr4_+_3768075 | 0.17 |
ENST00000509482.1
ENST00000330055.5 |
ADRA2C
|
adrenoceptor alpha 2C |
| chr11_+_66624527 | 0.16 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr1_+_162039558 | 0.16 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr8_+_145582231 | 0.16 |
ENST00000526338.1
ENST00000402965.1 ENST00000534725.1 ENST00000532887.1 ENST00000329994.2 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr22_-_50699972 | 0.16 |
ENST00000395778.3
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr11_-_842509 | 0.16 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr19_+_9945962 | 0.16 |
ENST00000587625.1
ENST00000247970.4 ENST00000588695.1 |
PIN1
|
peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
| chr18_+_77439775 | 0.16 |
ENST00000299543.7
ENST00000075430.7 |
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr1_-_1850697 | 0.16 |
ENST00000378598.4
ENST00000416272.1 ENST00000310991.3 |
TMEM52
|
transmembrane protein 52 |
| chr16_+_50280020 | 0.16 |
ENST00000564965.1
|
ADCY7
|
adenylate cyclase 7 |
| chr14_+_92980111 | 0.16 |
ENST00000216487.7
ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr12_+_26274917 | 0.16 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr5_+_149865377 | 0.16 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chrX_+_153686614 | 0.16 |
ENST00000369682.3
|
PLXNA3
|
plexin A3 |
| chr11_-_9482010 | 0.16 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr16_+_77233294 | 0.16 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr10_+_43916052 | 0.16 |
ENST00000442526.2
|
RP11-517P14.2
|
RP11-517P14.2 |
| chr16_-_80838160 | 0.16 |
ENST00000562812.1
ENST00000563890.1 ENST00000566173.1 |
CDYL2
|
chromodomain protein, Y-like 2 |
| chr12_-_90103077 | 0.16 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr16_-_4664382 | 0.16 |
ENST00000591113.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr1_-_84543614 | 0.15 |
ENST00000605506.1
|
RP11-486G15.2
|
RP11-486G15.2 |
| chr8_-_145742862 | 0.15 |
ENST00000524998.1
|
RECQL4
|
RecQ protein-like 4 |
| chr8_-_145013711 | 0.15 |
ENST00000345136.3
|
PLEC
|
plectin |
| chr9_+_131580734 | 0.15 |
ENST00000372642.4
|
ENDOG
|
endonuclease G |
| chr8_-_145582118 | 0.15 |
ENST00000455319.2
ENST00000331890.5 |
FBXL6
|
F-box and leucine-rich repeat protein 6 |
| chr9_-_140009130 | 0.15 |
ENST00000497375.1
ENST00000371579.2 |
DPP7
|
dipeptidyl-peptidase 7 |
| chr20_-_48532046 | 0.15 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr1_-_21948906 | 0.15 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr7_-_124405681 | 0.15 |
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr19_-_15543368 | 0.15 |
ENST00000599686.3
|
WIZ
|
widely interspaced zinc finger motifs |
| chr3_+_32148106 | 0.15 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr16_+_85646763 | 0.15 |
ENST00000411612.1
ENST00000253458.7 |
GSE1
|
Gse1 coiled-coil protein |
| chr19_-_2236246 | 0.15 |
ENST00000587394.2
|
PLEKHJ1
|
pleckstrin homology domain containing, family J member 1 |
| chr19_+_709101 | 0.15 |
ENST00000338448.5
|
PALM
|
paralemmin |
| chr18_+_77623668 | 0.15 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr22_+_37956453 | 0.15 |
ENST00000249014.4
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr13_-_36788718 | 0.15 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr17_+_78075361 | 0.15 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr13_+_98795664 | 0.15 |
ENST00000376581.5
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr6_+_139349903 | 0.15 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr2_-_139259244 | 0.14 |
ENST00000414911.1
ENST00000431985.1 |
AC097721.2
|
AC097721.2 |
| chr4_+_75023816 | 0.14 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr9_-_91793675 | 0.14 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr6_-_105584560 | 0.14 |
ENST00000336775.5
|
BVES
|
blood vessel epicardial substance |
| chr10_+_102759545 | 0.14 |
ENST00000454422.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr5_+_133450365 | 0.14 |
ENST00000342854.5
ENST00000321603.6 ENST00000321584.4 ENST00000378564.1 ENST00000395029.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr12_+_70760056 | 0.14 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr16_-_4664860 | 0.14 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr3_+_50192833 | 0.14 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr11_+_842808 | 0.14 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr16_-_79633799 | 0.14 |
ENST00000569649.1
|
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr14_+_96343100 | 0.14 |
ENST00000503525.2
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr20_+_49411523 | 0.14 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr6_+_1389989 | 0.14 |
ENST00000259806.1
|
FOXF2
|
forkhead box F2 |
| chr6_+_56820152 | 0.14 |
ENST00000370745.1
|
BEND6
|
BEN domain containing 6 |
| chr11_-_2292182 | 0.14 |
ENST00000331289.4
|
ASCL2
|
achaete-scute family bHLH transcription factor 2 |
| chr12_-_133464118 | 0.13 |
ENST00000540963.1
|
CHFR
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr11_+_64863587 | 0.13 |
ENST00000530773.1
ENST00000279281.3 ENST00000529180.1 |
VPS51
|
vacuolar protein sorting 51 homolog (S. cerevisiae) |
| chr16_+_838614 | 0.13 |
ENST00000262315.9
ENST00000455171.2 |
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr17_+_1627834 | 0.13 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr3_-_138048682 | 0.13 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr9_+_130922537 | 0.13 |
ENST00000372994.1
|
C9orf16
|
chromosome 9 open reading frame 16 |
| chr19_+_1286097 | 0.13 |
ENST00000215368.2
|
EFNA2
|
ephrin-A2 |
| chr13_+_110959598 | 0.13 |
ENST00000360467.5
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr5_-_175843524 | 0.13 |
ENST00000502877.1
|
CLTB
|
clathrin, light chain B |
| chr15_+_75640068 | 0.13 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr9_-_130742792 | 0.13 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr22_-_31741757 | 0.13 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr1_-_1284730 | 0.13 |
ENST00000378888.5
|
DVL1
|
dishevelled segment polarity protein 1 |
| chr17_-_15902951 | 0.13 |
ENST00000472495.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr14_-_51297360 | 0.13 |
ENST00000496749.1
|
NIN
|
ninein (GSK3B interacting protein) |
| chr22_+_37956479 | 0.13 |
ENST00000430687.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr2_+_23608064 | 0.13 |
ENST00000486442.1
|
KLHL29
|
kelch-like family member 29 |
| chr3_-_156534754 | 0.13 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr22_+_50609150 | 0.13 |
ENST00000159647.5
ENST00000395842.2 |
PANX2
|
pannexin 2 |
| chr12_-_6756559 | 0.13 |
ENST00000536350.1
ENST00000414226.2 ENST00000546114.1 |
ACRBP
|
acrosin binding protein |
| chrX_-_153363125 | 0.13 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr17_+_17685422 | 0.13 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr2_+_105760549 | 0.13 |
ENST00000436841.1
ENST00000452037.1 ENST00000438148.1 |
AC104655.3
|
AC104655.3 |
| chr11_-_65325430 | 0.12 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr11_-_60719213 | 0.12 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr16_+_765092 | 0.12 |
ENST00000568223.2
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr9_-_123639445 | 0.12 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr10_-_17659234 | 0.12 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr22_+_24820341 | 0.12 |
ENST00000464977.1
ENST00000444262.2 |
ADORA2A
|
adenosine A2a receptor |
| chr11_-_67272794 | 0.12 |
ENST00000436757.2
ENST00000356404.3 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr15_-_65579177 | 0.12 |
ENST00000444347.2
ENST00000261888.6 |
PARP16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr16_+_78133536 | 0.12 |
ENST00000402655.2
ENST00000406884.2 ENST00000539474.2 ENST00000569818.1 ENST00000355860.3 ENST00000408984.3 |
WWOX
|
WW domain containing oxidoreductase |
| chr10_-_17659357 | 0.12 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr8_+_144766617 | 0.12 |
ENST00000454097.1
ENST00000534303.1 ENST00000529833.1 ENST00000530574.1 ENST00000442058.2 ENST00000358656.4 ENST00000526970.1 ENST00000532158.1 |
ZNF707
|
zinc finger protein 707 |
| chr3_+_187871659 | 0.12 |
ENST00000416784.1
ENST00000430340.1 ENST00000414139.1 ENST00000454789.1 |
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr19_+_42580274 | 0.12 |
ENST00000359044.4
|
ZNF574
|
zinc finger protein 574 |
| chr16_-_58034357 | 0.12 |
ENST00000562909.1
|
ZNF319
|
zinc finger protein 319 |
| chr16_-_686235 | 0.12 |
ENST00000568773.1
ENST00000565163.1 ENST00000397665.2 ENST00000397666.2 ENST00000301686.8 ENST00000338401.4 ENST00000397664.4 ENST00000568830.1 |
C16orf13
|
chromosome 16 open reading frame 13 |
| chr7_-_3083472 | 0.12 |
ENST00000356408.3
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr17_+_19437132 | 0.12 |
ENST00000436810.2
ENST00000270570.4 ENST00000457293.1 ENST00000542886.1 ENST00000575023.1 ENST00000395585.1 |
SLC47A1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr1_-_1535455 | 0.12 |
ENST00000422725.1
|
C1orf233
|
chromosome 1 open reading frame 233 |
| chr9_+_136223414 | 0.12 |
ENST00000371964.4
|
SURF2
|
surfeit 2 |
| chr16_-_2185899 | 0.12 |
ENST00000262304.4
ENST00000423118.1 |
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr17_+_78075324 | 0.12 |
ENST00000570803.1
|
GAA
|
glucosidase, alpha; acid |
| chr12_-_53574376 | 0.12 |
ENST00000267085.4
ENST00000379850.3 ENST00000379846.1 ENST00000424990.1 |
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr19_+_18530184 | 0.12 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr14_+_62331592 | 0.12 |
ENST00000554436.1
|
CTD-2277K2.1
|
CTD-2277K2.1 |
| chr6_+_292253 | 0.12 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chrX_-_54209640 | 0.12 |
ENST00000375180.2
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr16_-_2264779 | 0.12 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr21_-_46359760 | 0.12 |
ENST00000330551.3
ENST00000397841.1 ENST00000380070.4 |
C21orf67
|
chromosome 21 open reading frame 67 |
| chr17_-_79481666 | 0.11 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr21_+_46825032 | 0.11 |
ENST00000400337.2
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr5_+_111755280 | 0.11 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr8_-_19614810 | 0.11 |
ENST00000524213.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr19_+_2236509 | 0.11 |
ENST00000221494.5
|
SF3A2
|
splicing factor 3a, subunit 2, 66kDa |
| chr8_+_99956759 | 0.11 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr17_+_78075498 | 0.11 |
ENST00000302262.3
|
GAA
|
glucosidase, alpha; acid |
| chr22_-_50746027 | 0.11 |
ENST00000425954.1
ENST00000449103.1 |
PLXNB2
|
plexin B2 |
| chr16_+_2076869 | 0.11 |
ENST00000424542.2
ENST00000432365.2 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr2_-_72375167 | 0.11 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr15_-_76352069 | 0.11 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr19_-_47975106 | 0.11 |
ENST00000539381.1
ENST00000594353.1 ENST00000542837.1 |
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chrX_+_38420623 | 0.11 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr19_+_1285890 | 0.11 |
ENST00000344663.3
|
MUM1
|
melanoma associated antigen (mutated) 1 |
| chr8_+_145582217 | 0.11 |
ENST00000530047.1
ENST00000527078.1 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr3_-_49395705 | 0.11 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr14_-_105635090 | 0.11 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr16_+_1756162 | 0.11 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr16_+_89984287 | 0.11 |
ENST00000555147.1
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
| chr19_-_38747172 | 0.11 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr1_+_8378140 | 0.11 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr19_+_45844032 | 0.11 |
ENST00000589837.1
|
KLC3
|
kinesin light chain 3 |
| chr13_-_95364389 | 0.11 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr4_-_75024085 | 0.11 |
ENST00000600169.1
|
AC093677.1
|
Uncharacterized protein |
| chr9_-_140353748 | 0.11 |
ENST00000371472.2
ENST00000371475.3 ENST00000265663.7 ENST00000437259.1 ENST00000392812.4 ENST00000371474.3 ENST00000371473.3 |
NSMF
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr19_-_58874112 | 0.11 |
ENST00000311044.3
ENST00000595763.1 ENST00000425453.3 |
ZNF497
|
zinc finger protein 497 |
| chr2_+_136499287 | 0.11 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr3_+_42544084 | 0.11 |
ENST00000543411.1
ENST00000438259.2 ENST00000439731.1 ENST00000325123.4 |
VIPR1
|
vasoactive intestinal peptide receptor 1 |
| chr7_-_74867509 | 0.11 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr2_+_235860690 | 0.11 |
ENST00000416021.1
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr1_+_205473865 | 0.11 |
ENST00000506215.1
ENST00000419301.1 |
CDK18
|
cyclin-dependent kinase 18 |
| chr8_-_145018905 | 0.11 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr1_-_54872059 | 0.11 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr11_-_62389449 | 0.11 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HIC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.4 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.4 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.3 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.3 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.2 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.1 | 0.2 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.1 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.2 | GO:0048242 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) epinephrine secretion(GO:0048242) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.2 | GO:1903947 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 0.2 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 0.3 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.1 | GO:0075528 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.2 | GO:0045402 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.0 | 0.1 | GO:2000683 | regulation of cellular response to X-ray(GO:2000683) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.2 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.4 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.4 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0014061 | regulation of norepinephrine secretion(GO:0014061) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.6 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0050668 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.0 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) response to erythropoietin(GO:0036017) cellular response to erythropoietin(GO:0036018) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.1 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.0 | GO:0022604 | regulation of cell morphogenesis(GO:0022604) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.0 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining(GO:2001033) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.2 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.3 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.0 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.3 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.0 | 0.0 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.7 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.0 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.4 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.2 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.3 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.2 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0042356 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0048244 | phytanoyl-CoA dioxygenase activity(GO:0048244) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0016418 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.3 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.0 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |