Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HESX1
Z-value: 0.45
Transcription factors associated with HESX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HESX1
|
ENSG00000163666.4 | HESX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HESX1 | hg19_v2_chr3_-_57260377_57260549 | -0.42 | 4.1e-01 | Click! |
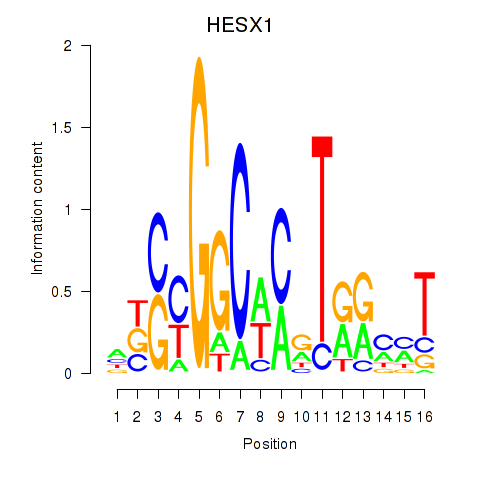
Activity profile of HESX1 motif
Sorted Z-values of HESX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_89952792 | 0.30 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr2_+_90458201 | 0.25 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr14_+_78227105 | 0.22 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr1_-_117021430 | 0.21 |
ENST00000423907.1
ENST00000434879.1 ENST00000443219.1 |
RP4-655J12.4
|
RP4-655J12.4 |
| chr19_+_45312310 | 0.19 |
ENST00000589651.1
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr7_+_4815238 | 0.18 |
ENST00000348624.4
ENST00000401897.1 |
AP5Z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chrX_-_84363974 | 0.17 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr10_-_5541525 | 0.17 |
ENST00000380332.3
|
CALML5
|
calmodulin-like 5 |
| chr21_-_38445443 | 0.15 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr1_-_20446020 | 0.14 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr19_+_45312347 | 0.14 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr12_-_752786 | 0.13 |
ENST00000433832.2
ENST00000542920.1 |
NINJ2
|
ninjurin 2 |
| chr2_+_32502952 | 0.13 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr12_+_65218352 | 0.13 |
ENST00000539867.1
ENST00000544457.1 ENST00000539120.1 |
TBC1D30
|
TBC1 domain family, member 30 |
| chr11_+_50257784 | 0.12 |
ENST00000529219.1
|
RP11-347H15.4
|
RP11-347H15.4 |
| chr19_-_45579762 | 0.11 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr6_-_41888843 | 0.11 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr17_+_73089382 | 0.11 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr3_-_196695733 | 0.11 |
ENST00000443835.1
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr19_-_19739321 | 0.11 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr5_-_88180342 | 0.11 |
ENST00000502983.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr4_+_160203650 | 0.11 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr3_-_51975942 | 0.10 |
ENST00000232888.6
|
RRP9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr1_+_11539204 | 0.10 |
ENST00000294484.6
ENST00000389575.3 |
PTCHD2
|
patched domain containing 2 |
| chr4_-_144826682 | 0.10 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr2_-_85828867 | 0.09 |
ENST00000425160.1
|
TMEM150A
|
transmembrane protein 150A |
| chr19_-_50370799 | 0.09 |
ENST00000600910.1
ENST00000322344.3 ENST00000600573.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr6_+_41888926 | 0.09 |
ENST00000230340.4
|
BYSL
|
bystin-like |
| chr12_+_10331605 | 0.09 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr17_+_19282064 | 0.09 |
ENST00000603493.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr19_-_39694894 | 0.08 |
ENST00000318438.6
|
SYCN
|
syncollin |
| chr7_-_2595337 | 0.08 |
ENST00000340611.4
|
BRAT1
|
BRCA1-associated ATM activator 1 |
| chr11_+_101918153 | 0.08 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr6_+_109416684 | 0.08 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr9_-_131486367 | 0.08 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr19_+_46498704 | 0.08 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr2_+_230787213 | 0.08 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr21_-_38445470 | 0.08 |
ENST00000399098.1
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_+_90248739 | 0.07 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr6_-_29600832 | 0.07 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr3_-_196695692 | 0.07 |
ENST00000412723.1
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr6_+_56820152 | 0.07 |
ENST00000370745.1
|
BEND6
|
BEN domain containing 6 |
| chr16_-_70719925 | 0.07 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr1_+_56046710 | 0.07 |
ENST00000422374.1
|
RP11-466L17.1
|
RP11-466L17.1 |
| chr1_-_1356628 | 0.07 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr20_+_1115821 | 0.06 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr15_-_89089860 | 0.06 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr2_+_85981008 | 0.06 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr17_-_36906058 | 0.06 |
ENST00000580830.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr3_-_46735132 | 0.06 |
ENST00000415953.1
|
ALS2CL
|
ALS2 C-terminal like |
| chr12_-_123717711 | 0.06 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr20_-_62587735 | 0.06 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr8_+_120885949 | 0.06 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr3_-_46735155 | 0.06 |
ENST00000318962.4
|
ALS2CL
|
ALS2 C-terminal like |
| chr20_-_40247002 | 0.06 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr20_+_18269121 | 0.06 |
ENST00000377671.3
ENST00000360010.5 ENST00000396026.3 ENST00000402618.2 ENST00000401790.1 ENST00000434018.1 ENST00000538547.1 ENST00000535822.1 |
ZNF133
|
zinc finger protein 133 |
| chr14_-_106068065 | 0.05 |
ENST00000390541.2
|
IGHE
|
immunoglobulin heavy constant epsilon |
| chrX_+_135730373 | 0.05 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr1_+_15256230 | 0.05 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr20_-_60982330 | 0.05 |
ENST00000279101.5
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2 |
| chr2_-_74875432 | 0.05 |
ENST00000536235.1
ENST00000421985.1 |
M1AP
|
meiosis 1 associated protein |
| chr7_-_72742085 | 0.05 |
ENST00000333149.2
|
TRIM50
|
tripartite motif containing 50 |
| chr6_+_26273144 | 0.05 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr12_+_53491220 | 0.05 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chrX_+_135730297 | 0.05 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr17_+_47572647 | 0.05 |
ENST00000172229.3
|
NGFR
|
nerve growth factor receptor |
| chr14_-_69262947 | 0.05 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr22_+_38321840 | 0.05 |
ENST00000454685.1
|
MICALL1
|
MICAL-like 1 |
| chr14_-_69262789 | 0.05 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr2_-_74726710 | 0.05 |
ENST00000377566.4
|
LBX2
|
ladybird homeobox 2 |
| chr4_-_2264015 | 0.04 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chrX_-_131623043 | 0.04 |
ENST00000421707.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr1_-_31661000 | 0.04 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr19_+_42388437 | 0.04 |
ENST00000378152.4
ENST00000337665.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr16_-_67217844 | 0.04 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr1_-_165738072 | 0.04 |
ENST00000481278.1
|
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr3_+_192958914 | 0.04 |
ENST00000264735.2
ENST00000602513.1 |
HRASLS
|
HRAS-like suppressor |
| chr19_+_2249308 | 0.04 |
ENST00000592877.1
ENST00000221496.4 |
AMH
|
anti-Mullerian hormone |
| chr16_+_56659687 | 0.04 |
ENST00000568293.1
ENST00000330439.6 |
MT1E
|
metallothionein 1E |
| chr12_-_11091862 | 0.04 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr12_+_123717967 | 0.04 |
ENST00000536130.1
ENST00000546132.1 |
C12orf65
|
chromosome 12 open reading frame 65 |
| chr16_-_66968055 | 0.04 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr16_+_1662326 | 0.04 |
ENST00000397412.3
|
CRAMP1L
|
Crm, cramped-like (Drosophila) |
| chr19_+_41698927 | 0.04 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr15_+_73976545 | 0.04 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr17_+_36861735 | 0.04 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr19_-_50370509 | 0.04 |
ENST00000596014.1
|
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr22_+_44351301 | 0.04 |
ENST00000350028.4
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr5_-_122759032 | 0.04 |
ENST00000510582.3
ENST00000328236.5 ENST00000306481.6 ENST00000508442.2 ENST00000395431.2 |
CEP120
|
centrosomal protein 120kDa |
| chr8_-_145060593 | 0.04 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr21_-_38445011 | 0.03 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr22_+_19419425 | 0.03 |
ENST00000333130.3
|
MRPL40
|
mitochondrial ribosomal protein L40 |
| chr22_+_20105259 | 0.03 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr19_+_13228917 | 0.03 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr19_+_41699135 | 0.03 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr17_+_46018872 | 0.03 |
ENST00000583599.1
ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr19_+_19496728 | 0.03 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr2_-_85829811 | 0.03 |
ENST00000306353.3
|
TMEM150A
|
transmembrane protein 150A |
| chr7_+_74508372 | 0.03 |
ENST00000356115.5
ENST00000430511.2 ENST00000312575.7 |
GTF2IRD2B
|
GTF2I repeat domain containing 2B |
| chr19_+_38397839 | 0.03 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr1_-_16302565 | 0.03 |
ENST00000537142.1
ENST00000448462.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr7_-_74267836 | 0.03 |
ENST00000361071.5
ENST00000453619.2 ENST00000417115.2 ENST00000405086.2 |
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr21_+_33784670 | 0.03 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr1_+_11796177 | 0.03 |
ENST00000400895.2
ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor-associated protein |
| chr22_+_18632666 | 0.03 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr11_-_71791435 | 0.03 |
ENST00000351960.6
ENST00000541719.1 ENST00000535111.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr1_+_3614591 | 0.03 |
ENST00000378290.4
|
TP73
|
tumor protein p73 |
| chr14_-_69262916 | 0.03 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr11_-_71791518 | 0.03 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr1_-_9953295 | 0.03 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr4_+_156588350 | 0.03 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr7_+_99746514 | 0.03 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr17_+_55163075 | 0.03 |
ENST00000571629.1
ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr8_+_97274119 | 0.03 |
ENST00000455950.2
|
PTDSS1
|
phosphatidylserine synthase 1 |
| chr18_-_70535177 | 0.02 |
ENST00000327305.6
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr9_+_37079968 | 0.02 |
ENST00000588403.1
|
RP11-220I1.1
|
RP11-220I1.1 |
| chr12_-_121019165 | 0.02 |
ENST00000341039.2
ENST00000357500.4 |
POP5
|
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr5_+_40679584 | 0.02 |
ENST00000302472.3
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr10_+_103348031 | 0.02 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr22_+_50354104 | 0.02 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr12_+_6644443 | 0.02 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr5_-_176057518 | 0.02 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr3_+_9944303 | 0.02 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr1_-_40237020 | 0.02 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr2_-_96931679 | 0.02 |
ENST00000258439.3
ENST00000432959.1 |
TMEM127
|
transmembrane protein 127 |
| chr11_+_73019282 | 0.02 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr12_-_123717643 | 0.02 |
ENST00000541437.1
ENST00000606320.1 |
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr6_-_144329384 | 0.02 |
ENST00000417959.2
|
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr1_-_26372602 | 0.02 |
ENST00000374278.3
ENST00000374276.3 |
SLC30A2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr9_+_132094579 | 0.02 |
ENST00000427109.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr1_+_11796126 | 0.01 |
ENST00000376637.3
|
AGTRAP
|
angiotensin II receptor-associated protein |
| chr13_-_52378231 | 0.01 |
ENST00000280056.2
ENST00000444610.2 |
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr8_+_27182862 | 0.01 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr19_-_19431298 | 0.01 |
ENST00000590439.2
ENST00000334782.5 |
SUGP1
|
SURP and G patch domain containing 1 |
| chr22_+_44351419 | 0.01 |
ENST00000396202.3
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr6_-_83775489 | 0.01 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr15_-_31453359 | 0.01 |
ENST00000542188.1
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr18_+_54318616 | 0.01 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr1_-_165738085 | 0.01 |
ENST00000464650.1
ENST00000392129.6 |
TMCO1
|
transmembrane and coiled-coil domains 1 |
| chr16_+_4784273 | 0.01 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr2_-_61765732 | 0.01 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr16_-_30134441 | 0.01 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr4_+_129349188 | 0.01 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr12_-_49259643 | 0.01 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr17_-_70053866 | 0.01 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr1_-_161337662 | 0.01 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr9_+_1051481 | 0.01 |
ENST00000358146.2
ENST00000259622.6 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr9_-_34376851 | 0.01 |
ENST00000297625.7
|
KIAA1161
|
KIAA1161 |
| chr2_-_70944855 | 0.01 |
ENST00000415348.1
|
ADD2
|
adducin 2 (beta) |
| chr5_-_171881491 | 0.01 |
ENST00000311601.5
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chrX_+_105969893 | 0.01 |
ENST00000255499.2
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr12_+_131438443 | 0.01 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr4_+_144303093 | 0.01 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr3_+_46921732 | 0.01 |
ENST00000418619.1
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr19_-_38397285 | 0.01 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chr12_-_112546547 | 0.01 |
ENST00000547133.1
|
NAA25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
| chr17_+_45286387 | 0.01 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr10_-_75634326 | 0.01 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr7_-_37024665 | 0.01 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr16_+_4784458 | 0.01 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr8_-_93029865 | 0.01 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_-_31138439 | 0.01 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr2_+_208576355 | 0.01 |
ENST00000420822.1
ENST00000295414.3 ENST00000339882.5 |
CCNYL1
|
cyclin Y-like 1 |
| chr20_+_35807512 | 0.01 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr20_+_47835884 | 0.01 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr10_+_90672113 | 0.01 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr20_-_5485166 | 0.01 |
ENST00000589201.1
ENST00000379053.4 |
LINC00654
|
long intergenic non-protein coding RNA 654 |
| chr18_+_54318566 | 0.01 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr5_-_159546396 | 0.01 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr17_-_32906388 | 0.01 |
ENST00000357754.1
|
C17orf102
|
chromosome 17 open reading frame 102 |
| chr9_+_37120536 | 0.01 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr1_-_204165610 | 0.01 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
| chr6_-_41888814 | 0.00 |
ENST00000409060.1
ENST00000265350.4 |
MED20
|
mediator complex subunit 20 |
| chr14_-_70883708 | 0.00 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr1_+_45265897 | 0.00 |
ENST00000372201.4
|
PLK3
|
polo-like kinase 3 |
| chr14_-_23395623 | 0.00 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr3_+_158519654 | 0.00 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr20_+_35807449 | 0.00 |
ENST00000237530.6
|
RPN2
|
ribophorin II |
| chr11_+_126139005 | 0.00 |
ENST00000263578.5
ENST00000442061.2 ENST00000532125.1 |
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr2_-_61765315 | 0.00 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr6_+_56820018 | 0.00 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr8_-_110656995 | 0.00 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr8_-_16035454 | 0.00 |
ENST00000355282.2
|
MSR1
|
macrophage scavenger receptor 1 |
| chr6_+_56819773 | 0.00 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr3_+_138153451 | 0.00 |
ENST00000389567.4
ENST00000289135.4 |
ESYT3
|
extended synaptotagmin-like protein 3 |
| chr22_+_38453207 | 0.00 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr8_-_97173020 | 0.00 |
ENST00000287020.5
|
GDF6
|
growth differentiation factor 6 |
| chr16_-_57219926 | 0.00 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr19_+_19496624 | 0.00 |
ENST00000494516.2
ENST00000360315.3 ENST00000252577.5 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr17_+_45286706 | 0.00 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr3_+_155860751 | 0.00 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr3_+_130648842 | 0.00 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr16_-_57219966 | 0.00 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of HESX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0070377 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.1 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.0 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.1 | 0.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |