Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
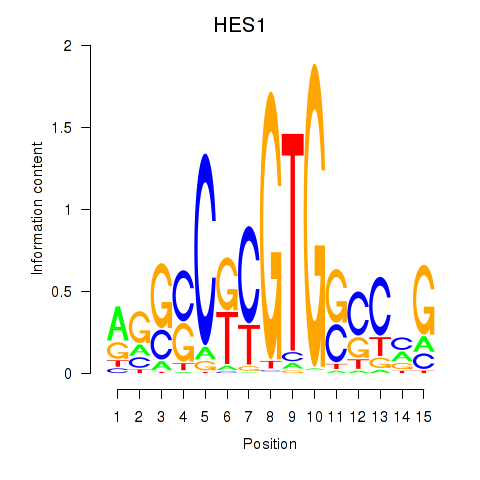
Results for HES1
Z-value: 0.96
Transcription factors associated with HES1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES1
|
ENSG00000114315.3 | hes family bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES1 | hg19_v2_chr3_+_193853927_193853944 | -0.22 | 6.8e-01 | Click! |
Activity profile of HES1 motif
Sorted Z-values of HES1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_9734004 | 0.45 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr6_-_30685214 | 0.39 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr14_+_94577074 | 0.38 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr8_-_142318398 | 0.37 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr21_+_34697258 | 0.36 |
ENST00000442071.1
ENST00000442357.2 |
IFNAR1
|
interferon (alpha, beta and omega) receptor 1 |
| chr11_+_64004888 | 0.35 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr19_+_54960790 | 0.34 |
ENST00000443957.1
|
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr5_+_667759 | 0.31 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr12_+_96588279 | 0.29 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr9_+_132427883 | 0.28 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chr20_+_6748311 | 0.28 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr11_-_111383064 | 0.28 |
ENST00000525791.1
ENST00000456861.2 ENST00000356018.2 |
BTG4
|
B-cell translocation gene 4 |
| chr17_-_57184260 | 0.27 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr2_-_86116093 | 0.27 |
ENST00000377332.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr18_-_72920372 | 0.26 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr8_+_25042192 | 0.26 |
ENST00000410074.1
|
DOCK5
|
dedicator of cytokinesis 5 |
| chr2_+_64681641 | 0.26 |
ENST00000409537.2
|
LGALSL
|
lectin, galactoside-binding-like |
| chr20_-_3154162 | 0.26 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr12_+_7022909 | 0.26 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr17_-_57184064 | 0.26 |
ENST00000262294.7
|
TRIM37
|
tripartite motif containing 37 |
| chr12_+_49761147 | 0.25 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr1_+_152881014 | 0.25 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr6_+_86159821 | 0.25 |
ENST00000369651.3
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr12_+_49760639 | 0.25 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr2_-_40006289 | 0.24 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chr9_+_101867387 | 0.23 |
ENST00000374990.2
ENST00000552516.1 |
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr17_-_57184170 | 0.23 |
ENST00000393065.2
|
TRIM37
|
tripartite motif containing 37 |
| chr1_+_44445549 | 0.23 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr5_-_7869108 | 0.22 |
ENST00000264669.5
ENST00000507572.1 ENST00000504695.1 |
FASTKD3
|
FAST kinase domains 3 |
| chr11_-_14521349 | 0.21 |
ENST00000534234.1
|
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr21_+_35445811 | 0.21 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr5_+_63461642 | 0.21 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr1_-_6761855 | 0.21 |
ENST00000426784.1
ENST00000377573.5 ENST00000377577.5 ENST00000294401.7 |
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11 |
| chr7_-_16460863 | 0.20 |
ENST00000407010.2
ENST00000399310.3 |
ISPD
|
isoprenoid synthase domain containing |
| chrX_+_21959108 | 0.20 |
ENST00000457085.1
|
SMS
|
spermine synthase |
| chr7_-_105925558 | 0.20 |
ENST00000222553.3
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr5_+_2752216 | 0.19 |
ENST00000457752.2
|
C5orf38
|
chromosome 5 open reading frame 38 |
| chr12_+_49658855 | 0.19 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr1_+_94884023 | 0.19 |
ENST00000315713.5
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr2_+_86116396 | 0.19 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chr12_+_107349606 | 0.19 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr18_-_33077868 | 0.19 |
ENST00000590757.1
ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C
RP11-322E11.6
|
INO80 complex subunit C Uncharacterized protein |
| chr18_-_33077556 | 0.19 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr2_-_224702201 | 0.19 |
ENST00000446015.2
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr1_-_38157877 | 0.18 |
ENST00000477060.1
ENST00000491981.1 ENST00000488137.1 |
C1orf109
|
chromosome 1 open reading frame 109 |
| chr16_-_4401284 | 0.18 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr18_-_77793891 | 0.18 |
ENST00000592957.1
ENST00000585474.1 |
TXNL4A
|
thioredoxin-like 4A |
| chr7_-_47621229 | 0.18 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr9_-_135545380 | 0.18 |
ENST00000544003.1
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr12_-_49525175 | 0.18 |
ENST00000336023.5
ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B
|
tubulin, alpha 1b |
| chr6_+_143771934 | 0.18 |
ENST00000367592.1
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr13_+_76123883 | 0.18 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr2_+_172949468 | 0.17 |
ENST00000361609.4
ENST00000469444.2 |
DLX1
|
distal-less homeobox 1 |
| chr4_-_76439483 | 0.17 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr12_+_107349497 | 0.17 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr17_-_48207115 | 0.17 |
ENST00000511964.1
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chr1_+_6845384 | 0.17 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr21_-_40685536 | 0.17 |
ENST00000341322.4
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr12_-_107487604 | 0.17 |
ENST00000008527.5
|
CRY1
|
cryptochrome 1 (photolyase-like) |
| chr1_+_153950202 | 0.17 |
ENST00000608236.1
|
RP11-422P24.11
|
RP11-422P24.11 |
| chr4_-_76439596 | 0.17 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr9_-_130889990 | 0.17 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr12_-_48213735 | 0.16 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr1_-_53793725 | 0.16 |
ENST00000371454.2
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr1_+_44445643 | 0.16 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr16_-_4466565 | 0.16 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr11_-_72852320 | 0.16 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr18_-_33077942 | 0.16 |
ENST00000334598.7
|
INO80C
|
INO80 complex subunit C |
| chr19_-_44124019 | 0.15 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr6_+_63921399 | 0.15 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr2_-_136743039 | 0.15 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr8_+_38614754 | 0.15 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr20_+_5986756 | 0.15 |
ENST00000452938.1
|
CRLS1
|
cardiolipin synthase 1 |
| chr15_-_49913126 | 0.15 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr11_-_615570 | 0.14 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr3_-_138763734 | 0.14 |
ENST00000413199.1
ENST00000502927.2 |
PRR23C
|
proline rich 23C |
| chr17_+_66031838 | 0.14 |
ENST00000584026.1
|
KPNA2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr15_-_44069741 | 0.14 |
ENST00000319359.3
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr2_-_224702270 | 0.14 |
ENST00000396654.2
ENST00000396653.2 ENST00000423110.1 ENST00000443700.1 |
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr15_+_41056218 | 0.14 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr19_+_39833036 | 0.14 |
ENST00000602243.1
ENST00000598913.1 ENST00000314471.6 |
SAMD4B
|
sterile alpha motif domain containing 4B |
| chr11_-_124632179 | 0.14 |
ENST00000278927.5
ENST00000442070.2 ENST00000444566.1 ENST00000435477.1 |
ESAM
|
endothelial cell adhesion molecule |
| chr17_+_26684604 | 0.14 |
ENST00000292114.3
ENST00000509083.1 |
TMEM199
|
transmembrane protein 199 |
| chr6_+_89790459 | 0.14 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr8_-_99954788 | 0.14 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr7_-_130597935 | 0.14 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr1_-_205290865 | 0.14 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr12_+_122064673 | 0.14 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr11_+_111896090 | 0.14 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr17_-_79849438 | 0.14 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr14_+_103243813 | 0.13 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr7_+_16685756 | 0.13 |
ENST00000415365.1
ENST00000258761.3 ENST00000433922.2 ENST00000452975.2 ENST00000405202.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr8_+_38614778 | 0.13 |
ENST00000521050.1
ENST00000522904.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr10_-_106240032 | 0.13 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr1_+_44444865 | 0.13 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr20_+_35201857 | 0.13 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr9_+_101867359 | 0.13 |
ENST00000374994.4
|
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr1_+_165797024 | 0.13 |
ENST00000372212.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr14_+_74004051 | 0.13 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr20_+_31407692 | 0.13 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr2_+_208576259 | 0.13 |
ENST00000392209.3
|
CCNYL1
|
cyclin Y-like 1 |
| chr12_+_49761273 | 0.13 |
ENST00000551540.1
ENST00000552918.1 ENST00000548777.1 ENST00000547865.1 ENST00000552171.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr19_+_49617581 | 0.13 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr22_+_46731596 | 0.13 |
ENST00000381019.3
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr17_-_1394940 | 0.13 |
ENST00000570984.2
ENST00000361007.2 |
MYO1C
|
myosin IC |
| chrX_+_24073048 | 0.13 |
ENST00000423068.1
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr6_-_143771799 | 0.12 |
ENST00000237283.8
|
ADAT2
|
adenosine deaminase, tRNA-specific 2 |
| chr15_+_89182178 | 0.12 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr22_-_24181174 | 0.12 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr12_+_7023491 | 0.12 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr21_-_18985230 | 0.12 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr5_-_54523143 | 0.12 |
ENST00000513312.1
|
MCIDAS
|
multiciliate differentiation and DNA synthesis associated cell cycle protein |
| chr9_-_38424443 | 0.12 |
ENST00000377694.1
|
IGFBPL1
|
insulin-like growth factor binding protein-like 1 |
| chr20_+_35201993 | 0.12 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr11_-_113746212 | 0.12 |
ENST00000537642.1
ENST00000537706.1 ENST00000544750.1 ENST00000260188.5 ENST00000540925.1 |
USP28
|
ubiquitin specific peptidase 28 |
| chr1_-_53793584 | 0.12 |
ENST00000354412.3
ENST00000347547.2 ENST00000306052.6 |
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr6_-_72129806 | 0.12 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr5_-_159797627 | 0.12 |
ENST00000393975.3
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2 |
| chr3_-_127541679 | 0.12 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr11_+_125757556 | 0.12 |
ENST00000526028.1
|
HYLS1
|
hydrolethalus syndrome 1 |
| chr13_+_37393351 | 0.12 |
ENST00000255476.2
|
RFXAP
|
regulatory factor X-associated protein |
| chr21_-_40685504 | 0.12 |
ENST00000380800.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr8_-_142011244 | 0.12 |
ENST00000340930.3
ENST00000520828.1 ENST00000524257.1 ENST00000523679.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr8_+_87526732 | 0.12 |
ENST00000523469.1
ENST00000522240.1 |
CPNE3
|
copine III |
| chr19_+_34891252 | 0.12 |
ENST00000606020.1
|
RP11-618P17.4
|
Uncharacterized protein |
| chr3_+_113667354 | 0.12 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr10_-_104001231 | 0.12 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr3_+_133292759 | 0.11 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr15_-_60771280 | 0.11 |
ENST00000560072.1
ENST00000560406.1 ENST00000560520.1 ENST00000261520.4 ENST00000439632.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr14_-_53162361 | 0.11 |
ENST00000395686.3
|
ERO1L
|
ERO1-like (S. cerevisiae) |
| chr1_+_6508571 | 0.11 |
ENST00000478323.1
|
ESPN
|
espin |
| chr15_-_67813924 | 0.11 |
ENST00000559298.1
|
IQCH-AS1
|
IQCH antisense RNA 1 |
| chr1_-_45452240 | 0.11 |
ENST00000372183.3
ENST00000372182.4 ENST00000360403.2 |
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa |
| chr2_+_150187020 | 0.11 |
ENST00000334166.4
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chr7_-_44365020 | 0.11 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr10_-_105110890 | 0.11 |
ENST00000369847.3
|
PCGF6
|
polycomb group ring finger 6 |
| chr1_-_118472171 | 0.11 |
ENST00000369442.3
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr17_+_77751931 | 0.11 |
ENST00000310942.4
ENST00000269399.5 |
CBX2
|
chromobox homolog 2 |
| chrX_-_54070388 | 0.11 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr7_+_7222157 | 0.11 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr17_+_7255208 | 0.11 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr6_+_86159765 | 0.11 |
ENST00000369646.3
ENST00000257770.3 |
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr12_-_92539614 | 0.11 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr17_-_47755338 | 0.11 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr11_+_65029233 | 0.11 |
ENST00000265465.3
|
POLA2
|
polymerase (DNA directed), alpha 2, accessory subunit |
| chr21_+_47878757 | 0.11 |
ENST00000400274.1
ENST00000427143.2 ENST00000318711.7 ENST00000457905.3 ENST00000466639.1 ENST00000435722.3 ENST00000417564.2 |
DIP2A
|
DIP2 disco-interacting protein 2 homolog A (Drosophila) |
| chr7_-_127032114 | 0.11 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chrX_-_20284958 | 0.11 |
ENST00000379565.3
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr11_+_12132117 | 0.11 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr9_-_88897426 | 0.11 |
ENST00000375991.4
ENST00000326094.4 |
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr1_+_41445413 | 0.11 |
ENST00000541520.1
|
CTPS1
|
CTP synthase 1 |
| chr22_+_38054721 | 0.11 |
ENST00000215904.6
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr9_+_33524240 | 0.11 |
ENST00000290943.6
|
ANKRD18B
|
ankyrin repeat domain 18B |
| chr1_-_8086343 | 0.11 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr18_+_77794446 | 0.10 |
ENST00000262197.7
|
RBFA
|
ribosome binding factor A (putative) |
| chr21_-_10990830 | 0.10 |
ENST00000361285.4
ENST00000342420.5 ENST00000328758.5 |
TPTE
|
transmembrane phosphatase with tensin homology |
| chr3_+_48956249 | 0.10 |
ENST00000452882.1
ENST00000430423.1 ENST00000356401.4 ENST00000449376.1 ENST00000420814.1 ENST00000449729.1 ENST00000433170.1 |
ARIH2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr1_+_225965518 | 0.10 |
ENST00000304786.7
ENST00000366839.4 ENST00000366838.1 |
SRP9
|
signal recognition particle 9kDa |
| chr1_-_38273840 | 0.10 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr16_+_66914264 | 0.10 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr2_-_10588630 | 0.10 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr19_+_33072373 | 0.10 |
ENST00000586035.1
|
PDCD5
|
programmed cell death 5 |
| chr10_+_90750378 | 0.10 |
ENST00000355740.2
ENST00000352159.4 |
FAS
|
Fas cell surface death receptor |
| chr1_-_32801825 | 0.10 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr9_+_133320301 | 0.10 |
ENST00000352480.5
|
ASS1
|
argininosuccinate synthase 1 |
| chr8_-_141645645 | 0.10 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr12_-_106641728 | 0.10 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr17_+_27052892 | 0.10 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr1_-_24306835 | 0.10 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr22_-_25801333 | 0.10 |
ENST00000444995.3
|
LRP5L
|
low density lipoprotein receptor-related protein 5-like |
| chr20_+_48807351 | 0.10 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr6_+_56954808 | 0.10 |
ENST00000510483.1
ENST00000370706.4 ENST00000357489.3 |
ZNF451
|
zinc finger protein 451 |
| chr11_-_10315741 | 0.10 |
ENST00000256190.8
|
SBF2
|
SET binding factor 2 |
| chr19_-_11669960 | 0.10 |
ENST00000589171.1
ENST00000590700.1 ENST00000586683.1 ENST00000593077.1 ENST00000252445.3 |
ELOF1
|
elongation factor 1 homolog (S. cerevisiae) |
| chr19_+_49622646 | 0.10 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr14_+_20937538 | 0.10 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr1_+_157963391 | 0.10 |
ENST00000359209.6
ENST00000416935.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr22_+_29168652 | 0.10 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr15_+_74908228 | 0.10 |
ENST00000566126.1
|
CLK3
|
CDC-like kinase 3 |
| chr13_+_21277482 | 0.10 |
ENST00000304920.3
|
IL17D
|
interleukin 17D |
| chr2_+_10091815 | 0.10 |
ENST00000324907.9
|
GRHL1
|
grainyhead-like 1 (Drosophila) |
| chr5_+_151151471 | 0.10 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr6_-_150346607 | 0.10 |
ENST00000367341.1
ENST00000286380.2 |
RAET1L
|
retinoic acid early transcript 1L |
| chr1_-_63153944 | 0.09 |
ENST00000340370.5
ENST00000404627.2 ENST00000251157.5 |
DOCK7
|
dedicator of cytokinesis 7 |
| chr16_+_2039946 | 0.09 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr7_-_155089251 | 0.09 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr2_+_139259324 | 0.09 |
ENST00000280098.4
|
SPOPL
|
speckle-type POZ protein-like |
| chr12_-_122238464 | 0.09 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr18_+_33767473 | 0.09 |
ENST00000261326.5
|
MOCOS
|
molybdenum cofactor sulfurase |
| chr3_+_182971583 | 0.09 |
ENST00000460419.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr12_+_1099675 | 0.09 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr2_-_62733476 | 0.09 |
ENST00000335390.5
|
TMEM17
|
transmembrane protein 17 |
| chr4_-_10459009 | 0.09 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr21_+_45527171 | 0.09 |
ENST00000291576.7
ENST00000456705.1 |
PWP2
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr8_-_94753229 | 0.09 |
ENST00000518597.1
ENST00000399300.2 ENST00000517700.1 |
RBM12B
|
RNA binding motif protein 12B |
| chr19_-_39390212 | 0.09 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr16_-_46865047 | 0.09 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr1_+_26438289 | 0.09 |
ENST00000374271.4
ENST00000374269.1 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chrX_-_48901012 | 0.09 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr12_-_89919965 | 0.09 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
Network of associatons between targets according to the STRING database.
First level regulatory network of HES1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.6 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 0.3 | GO:0060129 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) regulation of calcium-independent cell-cell adhesion(GO:0051040) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.4 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.4 | GO:1905071 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.4 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.2 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.2 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 0.3 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.0 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.1 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.0 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.0 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.0 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0072544 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.1 | 0.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.3 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 0.4 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.2 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.3 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.3 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.3 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |