Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
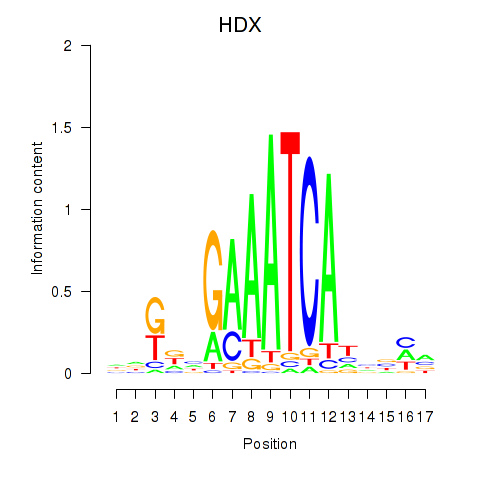
Results for HDX
Z-value: 0.46
Transcription factors associated with HDX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HDX
|
ENSG00000165259.9 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HDX | hg19_v2_chrX_-_83757399_83757487 | -0.67 | 1.5e-01 | Click! |
Activity profile of HDX motif
Sorted Z-values of HDX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_113735575 | 0.96 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr1_-_153363452 | 0.70 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr2_-_113594279 | 0.42 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr9_+_6215799 | 0.36 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr7_-_139756791 | 0.24 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr6_+_144471643 | 0.14 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr7_+_144015218 | 0.14 |
ENST00000408951.1
|
OR2A1
|
olfactory receptor, family 2, subfamily A, member 1 |
| chrX_+_100224676 | 0.12 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr14_+_71788096 | 0.12 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr19_+_36602104 | 0.11 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr5_-_135290705 | 0.10 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_-_209825674 | 0.08 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr7_-_107883678 | 0.08 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr11_-_66675371 | 0.07 |
ENST00000393955.2
|
PC
|
pyruvate carboxylase |
| chr8_+_24151620 | 0.07 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr1_-_169680745 | 0.06 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr9_+_35538616 | 0.06 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr13_+_24144796 | 0.06 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr3_-_100565249 | 0.05 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_+_117602925 | 0.05 |
ENST00000369466.4
|
TTF2
|
transcription termination factor, RNA polymerase II |
| chr3_-_51533966 | 0.05 |
ENST00000504652.1
|
VPRBP
|
Vpr (HIV-1) binding protein |
| chr7_-_55620433 | 0.05 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr12_+_101869096 | 0.05 |
ENST00000551346.1
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr1_+_21880560 | 0.05 |
ENST00000425315.2
|
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr1_-_11024258 | 0.04 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr14_+_79745682 | 0.04 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr5_-_135290651 | 0.04 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chrX_+_27608490 | 0.04 |
ENST00000451261.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr1_+_197237352 | 0.03 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr12_-_39734783 | 0.03 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr12_+_10460417 | 0.03 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr15_-_54025300 | 0.03 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr10_-_14372870 | 0.03 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chrX_+_55649833 | 0.03 |
ENST00000339140.3
|
FOXR2
|
forkhead box R2 |
| chr13_+_24144509 | 0.02 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr3_+_54157480 | 0.02 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr5_-_35230771 | 0.02 |
ENST00000342362.5
|
PRLR
|
prolactin receptor |
| chr2_+_128403439 | 0.02 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr8_+_110098850 | 0.02 |
ENST00000518632.1
|
TRHR
|
thyrotropin-releasing hormone receptor |
| chr7_+_99954224 | 0.02 |
ENST00000608825.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr7_-_121944491 | 0.01 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr7_-_135412925 | 0.01 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr14_+_79745746 | 0.00 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr18_-_53804580 | 0.00 |
ENST00000590484.1
ENST00000589293.1 ENST00000587904.1 ENST00000591974.1 |
RP11-456O19.4
|
RP11-456O19.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HDX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031622 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.1 | 0.7 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.4 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.0 | GO:0071529 | cementum mineralization(GO:0071529) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |