Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
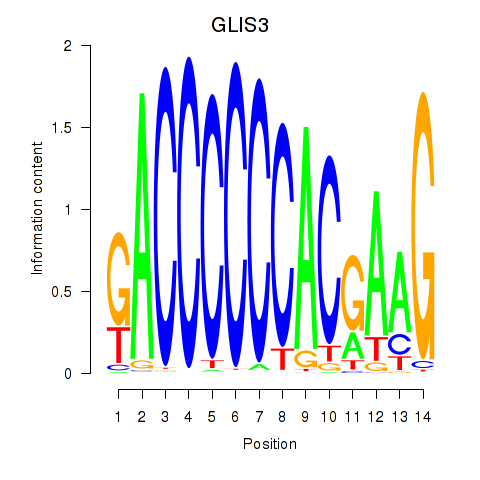
Results for GLIS3
Z-value: 0.27
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.17 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS3 | hg19_v2_chr9_-_4299874_4299916 | -0.00 | 1.0e+00 | Click! |
Activity profile of GLIS3 motif
Sorted Z-values of GLIS3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_65958693 | 0.22 |
ENST00000445681.1
ENST00000452565.1 |
GS1-124K5.4
|
GS1-124K5.4 |
| chr7_-_127032114 | 0.18 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr6_+_89791507 | 0.17 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr19_+_40697514 | 0.16 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr11_+_68451943 | 0.14 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chr12_+_132312931 | 0.13 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr22_+_50628999 | 0.10 |
ENST00000395827.1
|
TRABD
|
TraB domain containing |
| chr19_-_10613361 | 0.10 |
ENST00000591039.1
ENST00000591419.1 |
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr19_-_10613862 | 0.10 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr7_+_150758642 | 0.09 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr19_-_48673552 | 0.09 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr7_+_150758304 | 0.08 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr9_+_131903916 | 0.07 |
ENST00000419582.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_+_107599267 | 0.07 |
ENST00000361318.5
ENST00000370078.1 |
PRMT6
|
protein arginine methyltransferase 6 |
| chr1_+_31886653 | 0.07 |
ENST00000536384.1
|
SERINC2
|
serine incorporator 2 |
| chr12_+_48152774 | 0.07 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr1_+_59250815 | 0.07 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr9_+_131904233 | 0.06 |
ENST00000432651.1
ENST00000435132.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr19_+_41281060 | 0.06 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr11_+_393428 | 0.06 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr1_-_22215192 | 0.06 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr9_+_131904295 | 0.06 |
ENST00000434095.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr19_-_48673465 | 0.06 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr5_+_41925325 | 0.05 |
ENST00000296812.2
ENST00000281623.3 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr11_-_118789613 | 0.05 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chrX_-_132549506 | 0.05 |
ENST00000370828.3
|
GPC4
|
glypican 4 |
| chr22_-_31742218 | 0.04 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr6_+_44191290 | 0.04 |
ENST00000371755.3
ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr17_-_5015129 | 0.03 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr5_+_172386517 | 0.03 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chr5_-_180671172 | 0.03 |
ENST00000512805.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr22_-_50964849 | 0.03 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr19_-_41859814 | 0.03 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr19_+_13875316 | 0.03 |
ENST00000319545.8
ENST00000593245.1 ENST00000040663.6 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chr8_+_125860939 | 0.03 |
ENST00000525292.1
ENST00000528090.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr15_+_91478493 | 0.02 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr12_+_132568814 | 0.02 |
ENST00000454179.2
ENST00000389560.2 ENST00000443539.2 ENST00000392352.1 |
EP400NL
|
EP400 N-terminal like |
| chr19_-_49828438 | 0.02 |
ENST00000454748.3
ENST00000598828.1 ENST00000335875.4 |
SLC6A16
|
solute carrier family 6, member 16 |
| chr16_+_30006615 | 0.02 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr12_-_48152611 | 0.02 |
ENST00000389212.3
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr3_-_160167301 | 0.02 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chr11_+_68671310 | 0.02 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr6_+_44191507 | 0.02 |
ENST00000371724.1
ENST00000371713.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr22_-_21482352 | 0.02 |
ENST00000329949.3
|
POM121L7
|
POM121 transmembrane nucleoporin-like 7 |
| chr19_+_46850251 | 0.02 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr9_-_139658965 | 0.02 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr20_-_30310693 | 0.02 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr20_-_30310656 | 0.02 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr19_+_48828582 | 0.02 |
ENST00000270221.6
ENST00000596315.1 |
EMP3
|
epithelial membrane protein 3 |
| chr9_-_130742792 | 0.01 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr20_+_18447771 | 0.01 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr20_-_6103666 | 0.01 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr3_+_38029462 | 0.01 |
ENST00000283713.6
|
VILL
|
villin-like |
| chr20_-_44718538 | 0.01 |
ENST00000290231.6
ENST00000372291.3 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr12_-_57400227 | 0.01 |
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr15_-_43882353 | 0.01 |
ENST00000453080.1
ENST00000360301.4 ENST00000360135.4 ENST00000417085.1 ENST00000431962.1 ENST00000334933.4 ENST00000381879.4 ENST00000420765.1 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr14_+_77228532 | 0.01 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr15_-_43882140 | 0.01 |
ENST00000429176.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr6_+_33172407 | 0.01 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr5_-_137090028 | 0.01 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chrX_+_118370211 | 0.01 |
ENST00000217971.7
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr3_+_127348005 | 0.01 |
ENST00000342480.6
|
PODXL2
|
podocalyxin-like 2 |
| chr19_+_46850320 | 0.01 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr8_-_12051576 | 0.01 |
ENST00000524571.2
ENST00000533852.2 ENST00000533513.1 ENST00000448228.2 ENST00000534520.1 ENST00000321602.8 |
FAM86B1
|
family with sequence similarity 86, member B1 |
| chr8_+_118147498 | 0.01 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr3_-_160167508 | 0.01 |
ENST00000479460.1
|
TRIM59
|
tripartite motif containing 59 |
| chr1_-_157108266 | 0.01 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr20_+_61867235 | 0.00 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr2_+_220379052 | 0.00 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr7_+_23636992 | 0.00 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr9_-_98279241 | 0.00 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr7_-_50518022 | 0.00 |
ENST00000356889.4
ENST00000420829.1 ENST00000448788.1 ENST00000395556.2 ENST00000422854.1 ENST00000435566.1 ENST00000433017.1 |
FIGNL1
|
fidgetin-like 1 |
| chr17_-_73179046 | 0.00 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |