Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
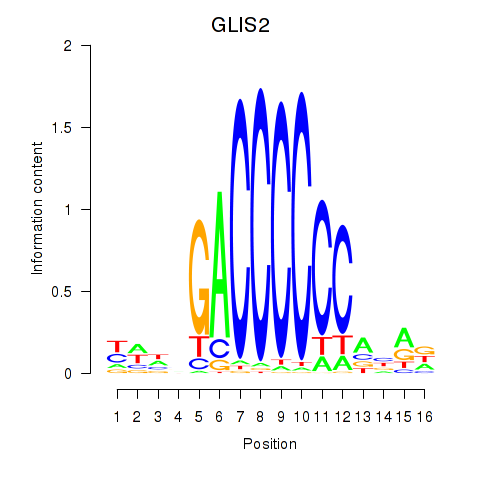
Results for GLIS2
Z-value: 1.31
Transcription factors associated with GLIS2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS2
|
ENSG00000126603.4 | GLIS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS2 | hg19_v2_chr16_+_4382225_4382225 | 0.78 | 6.8e-02 | Click! |
Activity profile of GLIS2 motif
Sorted Z-values of GLIS2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_44637526 | 1.20 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr11_+_844067 | 1.16 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr12_-_53242770 | 0.93 |
ENST00000304620.4
ENST00000547110.1 |
KRT78
|
keratin 78 |
| chr22_+_23264766 | 0.73 |
ENST00000390331.2
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr5_-_176836577 | 0.71 |
ENST00000253496.3
|
F12
|
coagulation factor XII (Hageman factor) |
| chr22_-_50699972 | 0.71 |
ENST00000395778.3
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr2_+_89952792 | 0.69 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr7_+_150758642 | 0.69 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr12_-_120907374 | 0.67 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr19_+_45254529 | 0.67 |
ENST00000444487.1
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr7_-_130080681 | 0.59 |
ENST00000469826.1
|
CEP41
|
centrosomal protein 41kDa |
| chr12_+_57998595 | 0.58 |
ENST00000337737.3
ENST00000548198.1 ENST00000551632.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr6_+_131571535 | 0.57 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr5_+_172484377 | 0.57 |
ENST00000523161.1
|
CREBRF
|
CREB3 regulatory factor |
| chr19_+_45417504 | 0.57 |
ENST00000588750.1
ENST00000588802.1 |
APOC1
|
apolipoprotein C-I |
| chr19_-_42931567 | 0.53 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr11_-_615942 | 0.52 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr11_+_66624527 | 0.51 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr7_-_73133959 | 0.51 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr2_+_37571717 | 0.49 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr14_+_105559784 | 0.49 |
ENST00000548104.1
|
RP11-44N21.1
|
RP11-44N21.1 |
| chr2_+_114737472 | 0.48 |
ENST00000420161.1
|
AC110769.3
|
AC110769.3 |
| chr11_-_795400 | 0.47 |
ENST00000526152.1
ENST00000456706.2 ENST00000528936.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr7_+_2685164 | 0.47 |
ENST00000400376.2
|
TTYH3
|
tweety family member 3 |
| chr19_-_19249255 | 0.47 |
ENST00000587583.2
ENST00000450333.2 ENST00000587096.1 ENST00000162044.9 ENST00000592369.1 ENST00000587915.1 |
TMEM161A
|
transmembrane protein 161A |
| chr7_+_76101379 | 0.46 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr19_-_17414179 | 0.46 |
ENST00000594194.1
ENST00000247706.3 |
ABHD8
|
abhydrolase domain containing 8 |
| chr19_-_36643329 | 0.44 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr19_+_17970693 | 0.43 |
ENST00000600147.1
ENST00000599898.1 |
RPL18A
|
ribosomal protein L18a |
| chrX_-_153744434 | 0.42 |
ENST00000369643.1
ENST00000393572.1 |
FAM3A
|
family with sequence similarity 3, member A |
| chr17_-_79481666 | 0.42 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr11_-_1619524 | 0.41 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr19_+_17970677 | 0.40 |
ENST00000222247.5
|
RPL18A
|
ribosomal protein L18a |
| chr11_-_615570 | 0.39 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr14_-_105635090 | 0.39 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr7_+_99746514 | 0.38 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr11_+_86748957 | 0.37 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chr19_+_41103063 | 0.36 |
ENST00000308370.7
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr7_+_2671663 | 0.36 |
ENST00000407643.1
|
TTYH3
|
tweety family member 3 |
| chr7_+_2695726 | 0.36 |
ENST00000429448.1
|
TTYH3
|
tweety family member 3 |
| chr19_-_8408139 | 0.34 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr11_-_795286 | 0.34 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr1_-_22215192 | 0.34 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr1_-_153348067 | 0.34 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr8_+_38243821 | 0.33 |
ENST00000519476.2
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr22_-_39640756 | 0.33 |
ENST00000331163.6
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr19_+_45349432 | 0.33 |
ENST00000252485.4
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr19_+_54369434 | 0.33 |
ENST00000421337.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr17_+_41158742 | 0.32 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr1_-_6550625 | 0.32 |
ENST00000377725.1
ENST00000340850.5 |
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr19_+_3506261 | 0.32 |
ENST00000441788.2
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila) |
| chr20_-_62203808 | 0.32 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr14_+_101293687 | 0.32 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_-_9938480 | 0.32 |
ENST00000585379.1
|
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr1_+_17866290 | 0.31 |
ENST00000361221.3
ENST00000452522.1 ENST00000434513.1 |
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr19_+_39926791 | 0.31 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr16_+_765092 | 0.31 |
ENST00000568223.2
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr22_-_50700140 | 0.31 |
ENST00000215659.8
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr7_+_100271446 | 0.31 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr19_-_6720686 | 0.30 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_+_3185910 | 0.30 |
ENST00000588428.1
|
NCLN
|
nicalin |
| chr11_+_64851729 | 0.30 |
ENST00000526791.1
ENST00000526945.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chrX_-_153744507 | 0.30 |
ENST00000442929.1
ENST00000426266.1 ENST00000359889.5 ENST00000369641.3 ENST00000447601.2 ENST00000434658.2 |
FAM3A
|
family with sequence similarity 3, member A |
| chr5_+_667759 | 0.29 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr2_+_37571845 | 0.29 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr9_+_139839686 | 0.29 |
ENST00000371634.2
|
C8G
|
complement component 8, gamma polypeptide |
| chr17_-_7120525 | 0.28 |
ENST00000447163.1
ENST00000399506.2 ENST00000302955.6 |
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr19_+_49661079 | 0.28 |
ENST00000355712.5
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr11_+_86748998 | 0.28 |
ENST00000525018.1
ENST00000355734.4 |
TMEM135
|
transmembrane protein 135 |
| chr11_+_393428 | 0.27 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr11_+_68451943 | 0.27 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chr16_-_18441131 | 0.27 |
ENST00000339303.5
|
NPIPA8
|
nuclear pore complex interacting protein family, member A8 |
| chr19_+_45417921 | 0.27 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr11_-_796197 | 0.27 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr3_+_127872265 | 0.27 |
ENST00000254730.6
ENST00000483457.1 |
EEFSEC
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr5_-_159739483 | 0.26 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr17_-_18907463 | 0.26 |
ENST00000585154.2
ENST00000345041.4 |
FAM83G
|
family with sequence similarity 83, member G |
| chr1_-_19229248 | 0.26 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr1_-_139379 | 0.26 |
ENST00000423372.3
|
AL627309.1
|
Uncharacterized protein |
| chr19_+_38397839 | 0.26 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr15_+_75498739 | 0.26 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr2_-_219154715 | 0.26 |
ENST00000451181.1
ENST00000429501.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr11_-_64646086 | 0.26 |
ENST00000320631.3
|
EHD1
|
EH-domain containing 1 |
| chr16_+_640201 | 0.25 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr19_+_45251804 | 0.25 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr19_+_45349630 | 0.25 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chrX_-_153602991 | 0.25 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr19_-_4902877 | 0.25 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr16_+_811073 | 0.25 |
ENST00000382862.3
ENST00000563651.1 |
MSLN
|
mesothelin |
| chr19_-_19006920 | 0.25 |
ENST00000429504.2
ENST00000427170.2 |
CERS1
|
ceramide synthase 1 |
| chr9_+_131903916 | 0.25 |
ENST00000419582.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr22_-_38484922 | 0.25 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr11_-_118789613 | 0.25 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr12_-_106480587 | 0.25 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr16_-_4664860 | 0.24 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr9_+_131904295 | 0.24 |
ENST00000434095.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr12_+_57854274 | 0.24 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr3_-_48470838 | 0.24 |
ENST00000358459.4
ENST00000358536.4 |
PLXNB1
|
plexin B1 |
| chr10_+_134232503 | 0.24 |
ENST00000428814.1
|
RP11-432J24.2
|
RP11-432J24.2 |
| chr19_+_49660997 | 0.23 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_+_17858509 | 0.23 |
ENST00000594202.1
ENST00000252771.7 ENST00000389133.4 |
FCHO1
|
FCH domain only 1 |
| chr10_-_102747190 | 0.23 |
ENST00000448244.1
ENST00000370241.3 ENST00000370236.1 ENST00000370234.4 ENST00000299179.5 ENST00000318325.2 ENST00000370242.4 ENST00000342071.1 ENST00000318364.8 ENST00000477279.1 |
MRPL43
|
mitochondrial ribosomal protein L43 |
| chr19_+_49622646 | 0.23 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr19_-_36236292 | 0.23 |
ENST00000378975.3
ENST00000412391.2 ENST00000292879.5 |
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr17_+_79953310 | 0.23 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr9_+_131904233 | 0.23 |
ENST00000432651.1
ENST00000435132.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr17_+_6939362 | 0.23 |
ENST00000308027.6
|
SLC16A13
|
solute carrier family 16, member 13 |
| chr9_+_97766469 | 0.23 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr11_+_832804 | 0.23 |
ENST00000397420.3
ENST00000525718.1 |
CD151
|
CD151 molecule (Raph blood group) |
| chr22_-_19974616 | 0.22 |
ENST00000344269.3
ENST00000401994.1 ENST00000406522.1 |
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome |
| chr1_+_2487800 | 0.22 |
ENST00000355716.4
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr22_+_31160239 | 0.22 |
ENST00000445781.1
ENST00000401475.1 |
OSBP2
|
oxysterol binding protein 2 |
| chr2_+_220492373 | 0.22 |
ENST00000317151.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr19_+_56159509 | 0.22 |
ENST00000586790.1
ENST00000591578.1 ENST00000588740.1 |
CCDC106
|
coiled-coil domain containing 106 |
| chr19_+_45417812 | 0.21 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr6_-_32908765 | 0.21 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr20_-_30310797 | 0.21 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr17_+_78075361 | 0.21 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr19_+_58111241 | 0.21 |
ENST00000597700.1
ENST00000332854.6 ENST00000597864.1 |
ZNF530
|
zinc finger protein 530 |
| chr19_+_10362882 | 0.21 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr19_-_2050852 | 0.21 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr16_+_84224740 | 0.21 |
ENST00000268624.3
|
ADAD2
|
adenosine deaminase domain containing 2 |
| chr22_-_18257178 | 0.20 |
ENST00000342111.5
|
BID
|
BH3 interacting domain death agonist |
| chr16_-_29934558 | 0.20 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr22_-_21482352 | 0.20 |
ENST00000329949.3
|
POM121L7
|
POM121 transmembrane nucleoporin-like 7 |
| chr19_-_10444188 | 0.20 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr11_+_64851666 | 0.20 |
ENST00000525509.1
ENST00000294258.3 ENST00000526334.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr11_+_3968573 | 0.20 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr19_+_11200038 | 0.20 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr19_+_56186557 | 0.19 |
ENST00000270460.6
|
EPN1
|
epsin 1 |
| chr19_-_41196534 | 0.19 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr1_-_1356628 | 0.19 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chrX_+_118108571 | 0.19 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr16_+_30087288 | 0.19 |
ENST00000279387.7
ENST00000562664.1 ENST00000562222.1 |
PPP4C
|
protein phosphatase 4, catalytic subunit |
| chr10_+_103825080 | 0.19 |
ENST00000299238.5
|
HPS6
|
Hermansky-Pudlak syndrome 6 |
| chr9_+_139839711 | 0.18 |
ENST00000224181.3
|
C8G
|
complement component 8, gamma polypeptide |
| chr22_+_21771656 | 0.18 |
ENST00000407464.2
|
HIC2
|
hypermethylated in cancer 2 |
| chr13_+_110959598 | 0.18 |
ENST00000360467.5
|
COL4A2
|
collagen, type IV, alpha 2 |
| chrX_+_53449887 | 0.18 |
ENST00000375327.3
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr19_+_3539152 | 0.18 |
ENST00000329493.5
|
C19orf71
|
chromosome 19 open reading frame 71 |
| chr4_-_111120334 | 0.18 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr19_-_18653781 | 0.18 |
ENST00000596558.2
ENST00000453489.2 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr22_-_46931191 | 0.18 |
ENST00000454637.1
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr12_+_122242597 | 0.18 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr16_-_29910365 | 0.17 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr19_+_56159421 | 0.17 |
ENST00000587213.1
|
CCDC106
|
coiled-coil domain containing 106 |
| chr16_-_89752965 | 0.17 |
ENST00000567544.1
|
RP11-368I7.4
|
Uncharacterized protein |
| chr16_-_2031464 | 0.17 |
ENST00000356120.4
ENST00000354249.4 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr9_+_117373486 | 0.17 |
ENST00000288502.4
ENST00000374049.4 |
C9orf91
|
chromosome 9 open reading frame 91 |
| chrX_-_153237258 | 0.17 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr9_-_34637806 | 0.16 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr17_+_38498594 | 0.16 |
ENST00000394081.3
|
RARA
|
retinoic acid receptor, alpha |
| chr10_+_94608245 | 0.16 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr17_-_79166176 | 0.16 |
ENST00000571292.1
|
AZI1
|
5-azacytidine induced 1 |
| chr3_-_16554403 | 0.16 |
ENST00000449415.1
ENST00000441460.1 |
RFTN1
|
raftlin, lipid raft linker 1 |
| chr19_+_14640372 | 0.16 |
ENST00000215567.5
ENST00000598298.1 ENST00000596073.1 ENST00000600083.1 ENST00000436007.2 |
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr19_+_1905365 | 0.16 |
ENST00000329478.2
ENST00000602400.1 ENST00000409472.1 |
ADAT3
SCAMP4
|
adenosine deaminase, tRNA-specific 3 secretory carrier membrane protein 4 |
| chr19_+_10362577 | 0.16 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chrX_-_102319092 | 0.16 |
ENST00000372728.3
|
BEX1
|
brain expressed, X-linked 1 |
| chr7_-_92107229 | 0.15 |
ENST00000603053.1
|
ERVW-1
|
endogenous retrovirus group W, member 1 |
| chr3_+_151531859 | 0.15 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr1_-_24127256 | 0.15 |
ENST00000418277.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr17_-_42200996 | 0.15 |
ENST00000587135.1
ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5
|
histone deacetylase 5 |
| chr3_-_193096600 | 0.15 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr22_+_21400229 | 0.15 |
ENST00000342608.4
ENST00000543388.1 ENST00000442047.1 |
AC002472.13
|
Leucine-rich repeat-containing protein LOC400891 |
| chr20_-_30310656 | 0.15 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr2_-_27718052 | 0.15 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr1_-_186649543 | 0.15 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr6_+_15249128 | 0.15 |
ENST00000397311.3
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr8_-_104427313 | 0.15 |
ENST00000297578.4
|
SLC25A32
|
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr19_-_41859814 | 0.15 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr19_-_12595586 | 0.15 |
ENST00000397732.3
|
ZNF709
|
zinc finger protein 709 |
| chr2_-_24346218 | 0.14 |
ENST00000436622.1
ENST00000313213.4 |
PFN4
|
profilin family, member 4 |
| chr17_-_27224621 | 0.14 |
ENST00000394906.2
ENST00000585169.1 ENST00000394908.4 |
FLOT2
|
flotillin 2 |
| chr10_+_134000404 | 0.14 |
ENST00000338492.4
ENST00000368629.1 |
DPYSL4
|
dihydropyrimidinase-like 4 |
| chr11_+_826136 | 0.14 |
ENST00000528315.1
ENST00000533803.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr17_-_4871085 | 0.14 |
ENST00000575142.1
ENST00000206020.3 |
SPAG7
|
sperm associated antigen 7 |
| chr22_-_30685596 | 0.14 |
ENST00000404953.3
ENST00000407689.3 |
GATSL3
|
GATS protein-like 3 |
| chr10_-_14880566 | 0.14 |
ENST00000378442.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr17_+_6658878 | 0.13 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr11_+_65292884 | 0.13 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr19_+_13875316 | 0.13 |
ENST00000319545.8
ENST00000593245.1 ENST00000040663.6 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chr5_-_141060389 | 0.13 |
ENST00000504448.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr14_-_21566731 | 0.13 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr19_+_56116771 | 0.13 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr19_+_17186577 | 0.13 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr19_-_3772209 | 0.12 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr17_-_5015129 | 0.12 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr20_+_48807351 | 0.12 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr12_+_57998400 | 0.12 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr6_-_32908792 | 0.12 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr19_+_45458503 | 0.12 |
ENST00000337392.5
ENST00000591304.1 |
CLPTM1
|
cleft lip and palate associated transmembrane protein 1 |
| chrX_+_54834159 | 0.12 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr19_-_17007026 | 0.12 |
ENST00000598792.1
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr8_-_11325047 | 0.12 |
ENST00000531804.1
|
FAM167A
|
family with sequence similarity 167, member A |
| chr4_-_111120132 | 0.12 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chrX_+_118108601 | 0.12 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr19_+_17858547 | 0.12 |
ENST00000600676.1
ENST00000600209.1 ENST00000596309.1 ENST00000598539.1 ENST00000597474.1 ENST00000593385.1 ENST00000598067.1 ENST00000593833.1 |
FCHO1
|
FCH domain only 1 |
| chr9_+_139847347 | 0.12 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr17_+_80416050 | 0.11 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.3 | 1.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.3 | 0.8 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.2 | 0.7 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 0.8 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 0.7 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.2 | 0.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.5 | GO:1990736 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 0.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.3 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.2 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.1 | 0.3 | GO:1990936 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.3 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.5 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0052510 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.3 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.0 | 1.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 1.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.4 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 1.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:1904298 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.0 | 0.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.1 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0060971 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.0 | GO:0072156 | distal tubule morphogenesis(GO:0072156) metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 0.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 1.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 1.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.3 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.1 | 0.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.2 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 2.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |