Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
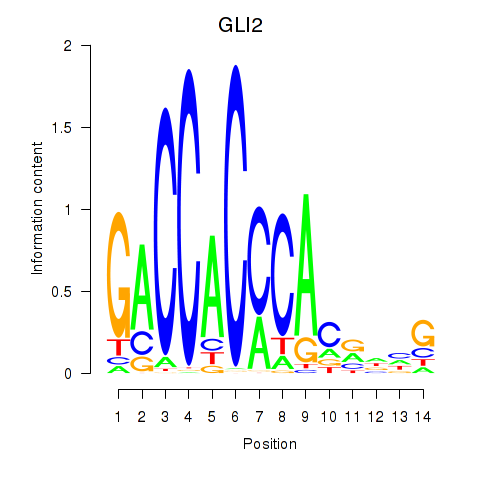
Results for GLI2
Z-value: 0.82
Transcription factors associated with GLI2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLI2
|
ENSG00000074047.16 | GLI family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI2 | hg19_v2_chr2_+_121493717_121493823 | -0.39 | 4.4e-01 | Click! |
Activity profile of GLI2 motif
Sorted Z-values of GLI2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_45417504 | 0.53 |
ENST00000588750.1
ENST00000588802.1 |
APOC1
|
apolipoprotein C-I |
| chr19_+_45417921 | 0.46 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr12_-_13529594 | 0.44 |
ENST00000539026.1
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr14_+_106938440 | 0.29 |
ENST00000433371.1
ENST00000449670.1 ENST00000334298.3 |
LINC00221
|
long intergenic non-protein coding RNA 221 |
| chr17_+_29248953 | 0.27 |
ENST00000581285.1
|
ADAP2
|
ArfGAP with dual PH domains 2 |
| chr2_-_197041044 | 0.26 |
ENST00000420683.1
|
STK17B
|
serine/threonine kinase 17b |
| chr8_+_96145974 | 0.26 |
ENST00000315367.3
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr11_-_111383064 | 0.26 |
ENST00000525791.1
ENST00000456861.2 ENST00000356018.2 |
BTG4
|
B-cell translocation gene 4 |
| chr19_-_40562063 | 0.24 |
ENST00000598845.1
ENST00000593605.1 ENST00000221355.6 ENST00000434248.1 |
ZNF780B
|
zinc finger protein 780B |
| chr16_-_30064244 | 0.24 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chr14_+_21467414 | 0.24 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr11_-_299519 | 0.24 |
ENST00000382614.2
|
IFITM5
|
interferon induced transmembrane protein 5 |
| chr18_-_59274139 | 0.23 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr6_-_121655850 | 0.23 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr12_+_132568814 | 0.22 |
ENST00000454179.2
ENST00000389560.2 ENST00000443539.2 ENST00000392352.1 |
EP400NL
|
EP400 N-terminal like |
| chr9_+_132427883 | 0.22 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chr8_+_74888417 | 0.21 |
ENST00000517439.1
ENST00000312184.5 |
TMEM70
|
transmembrane protein 70 |
| chr1_-_32403370 | 0.21 |
ENST00000534796.1
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr11_+_68451943 | 0.21 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chrX_-_135056106 | 0.21 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr7_+_142985467 | 0.21 |
ENST00000392925.2
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr14_+_105147464 | 0.21 |
ENST00000540171.2
|
RP11-982M15.6
|
RP11-982M15.6 |
| chr21_-_44035168 | 0.20 |
ENST00000419628.1
|
AP001626.1
|
AP001626.1 |
| chr12_-_110883346 | 0.19 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr11_-_73720276 | 0.19 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr17_+_29248918 | 0.19 |
ENST00000581548.1
ENST00000580525.1 |
ADAP2
|
ArfGAP with dual PH domains 2 |
| chr8_+_144371773 | 0.18 |
ENST00000523891.1
|
ZNF696
|
zinc finger protein 696 |
| chr6_+_26204825 | 0.18 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr19_-_44084586 | 0.18 |
ENST00000599693.1
ENST00000598165.1 |
XRCC1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr2_-_214017151 | 0.18 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr19_-_48673465 | 0.18 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chrX_+_153146127 | 0.18 |
ENST00000452593.1
ENST00000357566.1 |
LCA10
|
Putative lung carcinoma-associated protein 10 |
| chr7_-_127032363 | 0.18 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr17_-_76274572 | 0.18 |
ENST00000374945.1
|
RP11-219G17.4
|
RP11-219G17.4 |
| chr1_+_153600869 | 0.18 |
ENST00000292169.1
ENST00000368696.3 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chr1_+_228362251 | 0.18 |
ENST00000546123.1
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr22_+_23248512 | 0.17 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr16_+_2880254 | 0.17 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr16_-_71842706 | 0.17 |
ENST00000563104.1
ENST00000569975.1 ENST00000565412.1 ENST00000567583.1 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr11_-_111649015 | 0.17 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr1_+_152486950 | 0.17 |
ENST00000368790.3
|
CRCT1
|
cysteine-rich C-terminal 1 |
| chr22_-_46450024 | 0.17 |
ENST00000396008.2
ENST00000333761.1 |
C22orf26
|
chromosome 22 open reading frame 26 |
| chr6_-_53530474 | 0.17 |
ENST00000370905.3
|
KLHL31
|
kelch-like family member 31 |
| chr6_-_121655593 | 0.17 |
ENST00000398212.2
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr10_-_120938303 | 0.17 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr2_+_131369054 | 0.16 |
ENST00000409602.1
|
POTEJ
|
POTE ankyrin domain family, member J |
| chr7_-_960521 | 0.16 |
ENST00000437486.1
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr19_+_17858547 | 0.16 |
ENST00000600676.1
ENST00000600209.1 ENST00000596309.1 ENST00000598539.1 ENST00000597474.1 ENST00000593385.1 ENST00000598067.1 ENST00000593833.1 |
FCHO1
|
FCH domain only 1 |
| chr1_-_41487383 | 0.16 |
ENST00000302946.8
ENST00000372613.2 ENST00000439569.2 ENST00000397197.2 |
SLFNL1
|
schlafen-like 1 |
| chr17_+_1733251 | 0.16 |
ENST00000570451.1
|
RPA1
|
replication protein A1, 70kDa |
| chr1_-_21978312 | 0.16 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr1_+_6845578 | 0.16 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr3_+_11313995 | 0.16 |
ENST00000451513.1
ENST00000435760.1 ENST00000451830.1 |
ATG7
|
autophagy related 7 |
| chr11_-_73720122 | 0.15 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr9_-_35042824 | 0.15 |
ENST00000595331.1
|
FLJ00273
|
FLJ00273 |
| chr22_+_20008646 | 0.15 |
ENST00000401833.1
|
TANGO2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr6_-_121655552 | 0.15 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr16_-_15736345 | 0.15 |
ENST00000549219.1
|
KIAA0430
|
KIAA0430 |
| chr10_+_14880364 | 0.15 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr7_-_92465868 | 0.15 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr1_-_2343951 | 0.15 |
ENST00000288774.3
ENST00000507596.1 ENST00000447513.2 |
PEX10
|
peroxisomal biogenesis factor 10 |
| chr19_+_45417812 | 0.15 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr10_-_105156198 | 0.15 |
ENST00000369815.1
ENST00000309579.3 ENST00000337003.4 |
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr3_+_129207033 | 0.14 |
ENST00000507221.1
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr19_-_49176264 | 0.14 |
ENST00000270235.4
ENST00000596844.1 |
NTN5
|
netrin 5 |
| chr1_-_16563641 | 0.14 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr6_+_89790459 | 0.14 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr17_+_46185111 | 0.14 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr11_-_34938039 | 0.14 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr19_+_58962971 | 0.14 |
ENST00000336614.4
ENST00000545523.1 ENST00000599194.1 ENST00000598244.1 ENST00000599193.1 ENST00000594214.1 ENST00000391696.1 |
ZNF324B
|
zinc finger protein 324B |
| chr1_+_6845497 | 0.14 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr3_-_25824353 | 0.14 |
ENST00000427041.1
|
NGLY1
|
N-glycanase 1 |
| chr3_-_45957088 | 0.14 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr21_-_28215332 | 0.14 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr4_+_57843876 | 0.14 |
ENST00000450656.1
ENST00000381227.1 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr1_-_86861936 | 0.14 |
ENST00000394733.2
ENST00000359242.3 ENST00000294678.2 ENST00000479890.1 ENST00000317336.7 ENST00000370567.1 ENST00000394731.1 ENST00000478286.2 ENST00000370566.3 |
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr22_+_20008595 | 0.14 |
ENST00000398042.2
ENST00000450664.1 ENST00000327374.4 ENST00000432883.1 |
TANGO2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr11_-_31531121 | 0.14 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr21_-_40555393 | 0.14 |
ENST00000380900.2
|
PSMG1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr8_+_96146168 | 0.14 |
ENST00000519516.1
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr6_-_30685214 | 0.14 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr14_-_52535712 | 0.14 |
ENST00000216286.5
ENST00000541773.1 |
NID2
|
nidogen 2 (osteonidogen) |
| chr16_-_85146040 | 0.14 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr1_-_85358850 | 0.14 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr5_-_177659539 | 0.13 |
ENST00000476170.2
|
PHYKPL
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr8_-_86132568 | 0.13 |
ENST00000321777.5
ENST00000458398.2 ENST00000431163.2 ENST00000417663.2 ENST00000524353.1 ENST00000421308.2 ENST00000545322.1 ENST00000518562.1 |
C8orf59
|
chromosome 8 open reading frame 59 |
| chrX_+_70503526 | 0.13 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr2_+_183989157 | 0.13 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr11_-_118550346 | 0.13 |
ENST00000530256.1
|
TREH
|
trehalase (brush-border membrane glycoprotein) |
| chr1_+_14026671 | 0.13 |
ENST00000484063.2
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr22_+_23243156 | 0.13 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr22_-_50970506 | 0.13 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr17_-_30185946 | 0.13 |
ENST00000579741.1
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr10_+_6821545 | 0.13 |
ENST00000436383.1
|
LINC00707
|
long intergenic non-protein coding RNA 707 |
| chr11_-_63439013 | 0.13 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr3_-_155524049 | 0.13 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chrX_+_17755563 | 0.13 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr6_-_99963286 | 0.13 |
ENST00000369233.2
ENST00000500704.2 ENST00000329966.6 |
USP45
|
ubiquitin specific peptidase 45 |
| chr5_-_89705537 | 0.13 |
ENST00000522864.1
ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3
|
centrin, EF-hand protein, 3 |
| chr17_-_80059726 | 0.12 |
ENST00000583053.1
|
CCDC57
|
coiled-coil domain containing 57 |
| chr8_+_7705398 | 0.12 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr8_-_7320974 | 0.12 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr2_-_224702201 | 0.12 |
ENST00000446015.2
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr22_-_29137771 | 0.12 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr11_-_111649074 | 0.12 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_-_72496035 | 0.12 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr11_+_123301012 | 0.12 |
ENST00000533341.1
|
AP000783.1
|
Uncharacterized protein |
| chr17_+_80517216 | 0.12 |
ENST00000531030.1
ENST00000526383.2 |
FOXK2
|
forkhead box K2 |
| chr9_+_108456800 | 0.12 |
ENST00000434214.1
ENST00000374692.3 |
TMEM38B
|
transmembrane protein 38B |
| chr10_-_23633720 | 0.12 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr12_-_48099754 | 0.12 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr11_-_34937858 | 0.12 |
ENST00000278359.5
|
APIP
|
APAF1 interacting protein |
| chr5_-_95297534 | 0.12 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr17_-_60005365 | 0.12 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr19_-_49522727 | 0.12 |
ENST00000600007.1
|
CTB-60B18.10
|
CTB-60B18.10 |
| chr5_-_176923846 | 0.12 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr9_+_15553000 | 0.12 |
ENST00000297641.3
|
CCDC171
|
coiled-coil domain containing 171 |
| chr3_-_195538728 | 0.11 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr4_+_95128996 | 0.11 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr22_-_29138386 | 0.11 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr3_+_112930387 | 0.11 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chrX_-_30326445 | 0.11 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr11_-_71751715 | 0.11 |
ENST00000535947.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr19_+_56989485 | 0.11 |
ENST00000585445.1
ENST00000586091.1 ENST00000594783.1 ENST00000592146.1 ENST00000588158.1 ENST00000299997.4 ENST00000591797.1 |
ZNF667-AS1
|
ZNF667 antisense RNA 1 (head to head) |
| chr9_-_35071960 | 0.11 |
ENST00000417448.1
|
VCP
|
valosin containing protein |
| chr1_-_59012365 | 0.11 |
ENST00000456980.1
ENST00000482274.2 ENST00000453710.1 ENST00000419242.1 ENST00000358603.2 ENST00000371226.3 ENST00000426139.1 |
OMA1
|
OMA1 zinc metallopeptidase |
| chr19_-_23941639 | 0.11 |
ENST00000395385.3
ENST00000531570.1 ENST00000528059.1 |
ZNF681
|
zinc finger protein 681 |
| chr20_+_43514320 | 0.11 |
ENST00000372839.3
ENST00000428262.1 ENST00000445830.1 |
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
| chr10_-_127408011 | 0.11 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chr1_+_93646238 | 0.11 |
ENST00000448243.1
ENST00000370276.1 |
CCDC18
|
coiled-coil domain containing 18 |
| chr3_-_194207388 | 0.11 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr14_-_24615523 | 0.11 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr16_+_67063036 | 0.11 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr19_-_23941680 | 0.11 |
ENST00000402377.3
|
ZNF681
|
zinc finger protein 681 |
| chr6_+_26273144 | 0.11 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr15_-_75917955 | 0.11 |
ENST00000568162.1
ENST00000563875.1 |
SNUPN
|
snurportin 1 |
| chr14_-_75530693 | 0.11 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr17_-_30186328 | 0.11 |
ENST00000302362.6
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chrX_+_24073048 | 0.11 |
ENST00000423068.1
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr19_-_15343191 | 0.11 |
ENST00000221730.3
|
EPHX3
|
epoxide hydrolase 3 |
| chr15_-_78526942 | 0.11 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr2_+_183989083 | 0.11 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr7_+_30185496 | 0.11 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr11_+_3875757 | 0.11 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr19_-_10628098 | 0.11 |
ENST00000590601.1
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr5_-_82373260 | 0.11 |
ENST00000502346.1
|
TMEM167A
|
transmembrane protein 167A |
| chr11_-_118550375 | 0.11 |
ENST00000525958.1
ENST00000264029.4 ENST00000397925.1 ENST00000529101.1 |
TREH
|
trehalase (brush-border membrane glycoprotein) |
| chr11_+_31531291 | 0.11 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr22_+_37447771 | 0.11 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr17_+_18647326 | 0.11 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr22_+_39760130 | 0.11 |
ENST00000381535.4
|
SYNGR1
|
synaptogyrin 1 |
| chr5_-_156569754 | 0.10 |
ENST00000420343.1
|
MED7
|
mediator complex subunit 7 |
| chr19_+_37464063 | 0.10 |
ENST00000586353.1
ENST00000433993.1 ENST00000592567.1 |
ZNF568
|
zinc finger protein 568 |
| chr2_+_11295624 | 0.10 |
ENST00000402361.1
ENST00000428481.1 |
PQLC3
|
PQ loop repeat containing 3 |
| chr4_-_71705590 | 0.10 |
ENST00000254799.6
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr19_-_16284286 | 0.10 |
ENST00000379859.3
ENST00000269878.4 |
CIB3
|
calcium and integrin binding family member 3 |
| chr5_+_102455853 | 0.10 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr14_-_36983034 | 0.10 |
ENST00000518529.2
|
SFTA3
|
surfactant associated 3 |
| chr19_-_37263723 | 0.10 |
ENST00000589390.1
|
ZNF850
|
zinc finger protein 850 |
| chr9_-_139658965 | 0.10 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr5_+_172332220 | 0.10 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr22_+_39410287 | 0.10 |
ENST00000381568.4
|
APOBEC3D
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
| chrX_+_135618258 | 0.10 |
ENST00000440515.1
ENST00000456412.1 |
VGLL1
|
vestigial like 1 (Drosophila) |
| chr4_+_56814968 | 0.10 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr1_-_156269428 | 0.10 |
ENST00000339922.3
|
VHLL
|
von Hippel-Lindau tumor suppressor-like |
| chr1_+_225117350 | 0.10 |
ENST00000413949.2
ENST00000430092.1 ENST00000366850.3 ENST00000400952.3 ENST00000366849.1 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr12_-_108991812 | 0.10 |
ENST00000392806.3
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chr20_+_32782375 | 0.10 |
ENST00000568305.1
|
ASIP
|
agouti signaling protein |
| chr9_+_130860583 | 0.10 |
ENST00000373064.5
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr19_-_54567035 | 0.10 |
ENST00000366170.2
ENST00000425006.2 |
VSTM1
|
V-set and transmembrane domain containing 1 |
| chr1_+_162531294 | 0.10 |
ENST00000367926.4
ENST00000271469.3 |
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr1_+_154947126 | 0.10 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr4_-_156298087 | 0.10 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr5_-_89825328 | 0.10 |
ENST00000500869.2
ENST00000315948.6 ENST00000509384.1 |
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr13_-_52026730 | 0.10 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr16_-_3285144 | 0.10 |
ENST00000431561.3
ENST00000396870.4 |
ZNF200
|
zinc finger protein 200 |
| chr21_-_35340759 | 0.10 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr1_-_243417762 | 0.10 |
ENST00000522191.1
|
CEP170
|
centrosomal protein 170kDa |
| chr1_-_26232951 | 0.10 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr1_+_150522222 | 0.10 |
ENST00000369039.5
|
ADAMTSL4
|
ADAMTS-like 4 |
| chr9_+_78505581 | 0.10 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr4_-_156297919 | 0.10 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr4_-_156297949 | 0.09 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr12_-_13529642 | 0.09 |
ENST00000318426.2
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr4_-_107237374 | 0.09 |
ENST00000361687.4
ENST00000507696.1 ENST00000394708.2 ENST00000509532.1 |
TBCK
|
TBC1 domain containing kinase |
| chr6_-_99963252 | 0.09 |
ENST00000392738.2
ENST00000327681.6 ENST00000472914.2 |
USP45
|
ubiquitin specific peptidase 45 |
| chr3_-_25824925 | 0.09 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr10_-_14880002 | 0.09 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr2_+_219283815 | 0.09 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr22_+_29168652 | 0.09 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr20_+_44746939 | 0.09 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr18_+_12948000 | 0.09 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr12_+_120740119 | 0.09 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr12_-_48099773 | 0.09 |
ENST00000432584.3
ENST00000005386.3 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr8_-_128960591 | 0.09 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr3_+_156008809 | 0.09 |
ENST00000302490.8
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr17_-_71258019 | 0.09 |
ENST00000344935.4
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr3_-_23958506 | 0.09 |
ENST00000425478.2
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr7_-_112726393 | 0.09 |
ENST00000449591.1
ENST00000449735.1 ENST00000438062.1 ENST00000424100.1 |
GPR85
|
G protein-coupled receptor 85 |
| chr11_+_34938119 | 0.09 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLI2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.1 | 0.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.2 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.1 | 0.2 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.1 | 0.2 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.1 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.3 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.2 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.1 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.4 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0060971 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.0 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.4 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.1 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.0 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.1 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0036017 | response to erythropoietin(GO:0036017) cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.0 | GO:2000282 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) regulation of cellular amino acid biosynthetic process(GO:2000282) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0003335 | corneocyte development(GO:0003335) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.5 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.3 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.6 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.5 | GO:0003909 | DNA ligase activity(GO:0003909) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.0 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.0 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0052842 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |