Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
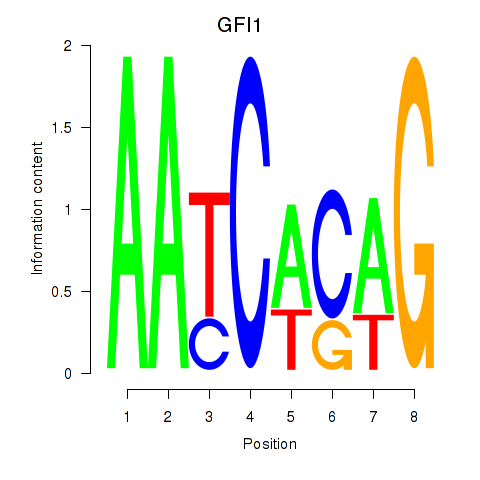
Results for GFI1
Z-value: 0.57
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1
|
ENSG00000162676.7 | growth factor independent 1 transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg19_v2_chr1_-_92951607_92951661 | -0.76 | 7.8e-02 | Click! |
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_56412713 | 0.54 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr2_-_111291587 | 0.40 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr15_+_96875657 | 0.29 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chrX_-_54069253 | 0.28 |
ENST00000425862.1
ENST00000433120.1 |
PHF8
|
PHD finger protein 8 |
| chr1_-_67142710 | 0.27 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr6_-_56492816 | 0.25 |
ENST00000522360.1
|
DST
|
dystonin |
| chr12_+_44229846 | 0.25 |
ENST00000551577.1
ENST00000266534.3 |
TMEM117
|
transmembrane protein 117 |
| chr6_-_52859046 | 0.25 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr1_+_154377669 | 0.24 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr11_+_10326918 | 0.23 |
ENST00000528544.1
|
ADM
|
adrenomedullin |
| chr14_-_106069247 | 0.23 |
ENST00000479229.1
|
RP11-731F5.1
|
RP11-731F5.1 |
| chr5_-_149465990 | 0.23 |
ENST00000543093.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr12_+_65996599 | 0.23 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr7_+_23637118 | 0.23 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr1_-_92764523 | 0.23 |
ENST00000370360.3
ENST00000534881.1 |
GLMN
|
glomulin, FKBP associated protein |
| chr5_+_118690466 | 0.21 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr7_+_23636992 | 0.21 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr11_-_94964354 | 0.21 |
ENST00000536441.1
|
SESN3
|
sestrin 3 |
| chr16_+_2533020 | 0.21 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chrX_-_15872914 | 0.21 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_-_99485825 | 0.20 |
ENST00000423771.1
|
KIAA1211L
|
KIAA1211-like |
| chr3_-_33686925 | 0.20 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr7_+_90339169 | 0.19 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr3_+_149689066 | 0.19 |
ENST00000593416.1
|
AC117395.1
|
LOC646903 protein; Uncharacterized protein |
| chr8_+_62200509 | 0.19 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr21_-_36421535 | 0.18 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr1_+_164528616 | 0.18 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr14_+_21525981 | 0.18 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chrX_+_100353153 | 0.18 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr15_+_96876340 | 0.17 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_+_8276433 | 0.17 |
ENST00000345999.3
ENST00000352620.3 ENST00000360500.3 |
CLEC4A
|
C-type lectin domain family 4, member A |
| chr4_-_185655212 | 0.17 |
ENST00000541971.1
|
MLF1IP
|
centromere protein U |
| chrX_+_154444643 | 0.17 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr8_-_123139423 | 0.17 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr9_+_2158485 | 0.17 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr13_-_27745936 | 0.17 |
ENST00000282344.6
|
USP12
|
ubiquitin specific peptidase 12 |
| chr4_-_157892055 | 0.16 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr7_-_84122033 | 0.16 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr19_-_49140692 | 0.16 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr21_-_36421626 | 0.16 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr10_-_65028817 | 0.16 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr12_-_31945172 | 0.15 |
ENST00000340398.3
|
H3F3C
|
H3 histone, family 3C |
| chr10_-_65028938 | 0.15 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr5_-_142000883 | 0.15 |
ENST00000359370.6
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr7_-_84121858 | 0.15 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_+_17721937 | 0.15 |
ENST00000451533.1
|
VSNL1
|
visinin-like 1 |
| chr4_+_86748898 | 0.15 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr9_+_2158443 | 0.15 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_-_155533787 | 0.14 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr3_+_149535022 | 0.14 |
ENST00000466795.1
|
RNF13
|
ring finger protein 13 |
| chr1_+_164528866 | 0.14 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr21_+_17214724 | 0.14 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr1_+_163039143 | 0.14 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr16_-_79634595 | 0.14 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr1_+_212475148 | 0.13 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr15_+_84116106 | 0.13 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr17_-_27467418 | 0.13 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr10_+_94451574 | 0.13 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr1_-_48866517 | 0.13 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr2_+_153574428 | 0.13 |
ENST00000326446.5
|
ARL6IP6
|
ADP-ribosylation-like factor 6 interacting protein 6 |
| chr7_+_43803790 | 0.13 |
ENST00000424330.1
|
BLVRA
|
biliverdin reductase A |
| chr21_-_36421401 | 0.13 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr11_-_82782861 | 0.13 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr1_+_23695680 | 0.13 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr5_+_133707252 | 0.13 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr4_-_70626430 | 0.13 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr6_-_41673552 | 0.12 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr19_+_41281060 | 0.12 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr8_+_132320792 | 0.12 |
ENST00000519695.1
ENST00000524275.1 |
CTD-2501M5.1
|
CTD-2501M5.1 |
| chr4_+_41614909 | 0.12 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_+_36393367 | 0.12 |
ENST00000246551.4
|
HCST
|
hematopoietic cell signal transducer |
| chr3_-_45957088 | 0.12 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr11_-_85780853 | 0.12 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_+_90192974 | 0.12 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr6_-_88411911 | 0.12 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr12_-_31158902 | 0.12 |
ENST00000544329.1
ENST00000418254.2 ENST00000222396.5 |
RP11-551L14.4
|
RP11-551L14.4 |
| chr13_-_30951282 | 0.12 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr2_+_103378472 | 0.12 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr15_-_70994612 | 0.12 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr5_+_59783941 | 0.12 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr2_+_158733088 | 0.12 |
ENST00000605860.1
|
UPP2
|
uridine phosphorylase 2 |
| chr19_+_48958766 | 0.11 |
ENST00000342291.2
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr7_+_102553430 | 0.11 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr9_-_75695323 | 0.11 |
ENST00000419959.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr19_-_40030861 | 0.11 |
ENST00000390658.2
|
EID2
|
EP300 interacting inhibitor of differentiation 2 |
| chr3_-_196910721 | 0.11 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr21_+_38593701 | 0.11 |
ENST00000440629.1
|
AP001432.14
|
AP001432.14 |
| chr2_+_66662690 | 0.11 |
ENST00000488550.1
|
MEIS1
|
Meis homeobox 1 |
| chr4_+_41937131 | 0.11 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr9_-_100459639 | 0.11 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr6_+_26440700 | 0.11 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr15_-_42448788 | 0.11 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr6_-_111804393 | 0.11 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr2_-_46844242 | 0.11 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr4_+_41362796 | 0.11 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_-_9092018 | 0.11 |
ENST00000397910.4
|
MUC16
|
mucin 16, cell surface associated |
| chr13_-_108867101 | 0.11 |
ENST00000356922.4
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chrX_-_138914394 | 0.11 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr4_-_84376953 | 0.11 |
ENST00000510985.1
ENST00000295488.3 |
HELQ
|
helicase, POLQ-like |
| chr3_-_45957534 | 0.11 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr6_-_143771799 | 0.11 |
ENST00000237283.8
|
ADAT2
|
adenosine deaminase, tRNA-specific 2 |
| chr5_-_139930713 | 0.11 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chr2_-_152830441 | 0.10 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_+_79783788 | 0.10 |
ENST00000282226.4
|
FAM151B
|
family with sequence similarity 151, member B |
| chr7_+_151791095 | 0.10 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr11_+_34642656 | 0.10 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr15_+_49715449 | 0.10 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr1_-_184943610 | 0.10 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr11_+_71791359 | 0.10 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr7_+_120591170 | 0.10 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr20_-_1569278 | 0.10 |
ENST00000262929.5
ENST00000567028.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr2_+_62900986 | 0.10 |
ENST00000405015.3
ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1
|
EH domain binding protein 1 |
| chr11_-_85430356 | 0.10 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr10_+_64564469 | 0.10 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr7_-_111428957 | 0.10 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr7_-_38948774 | 0.10 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr1_-_74663825 | 0.10 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr19_-_49140609 | 0.10 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr7_-_81635106 | 0.10 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr4_+_89300158 | 0.10 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr6_-_146285221 | 0.10 |
ENST00000367503.3
ENST00000438092.2 ENST00000275233.7 |
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr14_-_95623607 | 0.10 |
ENST00000531162.1
ENST00000529720.1 ENST00000343455.3 |
DICER1
|
dicer 1, ribonuclease type III |
| chr1_+_236557569 | 0.10 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr12_-_42878101 | 0.10 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr5_+_41925325 | 0.10 |
ENST00000296812.2
ENST00000281623.3 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr5_-_111091948 | 0.09 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr20_+_23168759 | 0.09 |
ENST00000411595.1
|
RP4-737E23.2
|
RP4-737E23.2 |
| chr17_+_27573875 | 0.09 |
ENST00000225387.3
|
CRYBA1
|
crystallin, beta A1 |
| chr14_-_64108537 | 0.09 |
ENST00000554717.1
ENST00000394942.2 |
WDR89
|
WD repeat domain 89 |
| chr5_+_109025067 | 0.09 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr5_+_157158205 | 0.09 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr11_+_85956182 | 0.09 |
ENST00000327320.4
ENST00000351625.6 ENST00000534595.1 |
EED
|
embryonic ectoderm development |
| chr3_-_142166904 | 0.09 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr19_-_57678811 | 0.09 |
ENST00000554048.2
|
DUXA
|
double homeobox A |
| chr19_+_47813110 | 0.09 |
ENST00000355085.3
|
C5AR1
|
complement component 5a receptor 1 |
| chr7_-_92146729 | 0.09 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr6_+_80816372 | 0.09 |
ENST00000545529.1
|
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr11_+_71791849 | 0.09 |
ENST00000423494.2
ENST00000539587.1 ENST00000538478.1 ENST00000324866.7 ENST00000439209.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr19_+_44488330 | 0.09 |
ENST00000591532.1
ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155
|
zinc finger protein 155 |
| chr7_-_151217166 | 0.09 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr3_+_141596371 | 0.09 |
ENST00000495216.1
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr1_-_114414316 | 0.09 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr6_+_111195973 | 0.09 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr10_-_14996070 | 0.09 |
ENST00000378258.1
ENST00000453695.2 ENST00000378246.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr12_+_26126681 | 0.09 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_-_150669500 | 0.09 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr1_-_152196669 | 0.09 |
ENST00000368801.2
|
HRNR
|
hornerin |
| chr3_-_48632593 | 0.08 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr11_-_299519 | 0.08 |
ENST00000382614.2
|
IFITM5
|
interferon induced transmembrane protein 5 |
| chr2_+_46844290 | 0.08 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr7_-_102985035 | 0.08 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr14_+_75179840 | 0.08 |
ENST00000554590.1
ENST00000341162.4 ENST00000534938.2 ENST00000553615.1 |
FCF1
|
FCF1 rRNA-processing protein |
| chr3_-_178976996 | 0.08 |
ENST00000485523.1
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr1_+_164529004 | 0.08 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_62439037 | 0.08 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_+_196621002 | 0.08 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr2_+_210443993 | 0.08 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr17_-_57229155 | 0.08 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_+_74663896 | 0.08 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chr3_-_188665428 | 0.08 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr12_+_110718428 | 0.08 |
ENST00000552636.1
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr2_+_87755054 | 0.08 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr11_+_10326612 | 0.08 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr2_+_47630108 | 0.08 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr9_+_2159850 | 0.08 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_+_48807155 | 0.08 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr16_+_21623958 | 0.08 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr3_-_168864315 | 0.08 |
ENST00000475754.1
ENST00000484519.1 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr22_-_28315115 | 0.08 |
ENST00000455418.3
ENST00000436663.1 ENST00000320996.10 ENST00000335272.5 |
PITPNB
|
phosphatidylinositol transfer protein, beta |
| chr1_+_156338993 | 0.08 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chrX_-_41782683 | 0.08 |
ENST00000378163.1
ENST00000378154.1 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr16_+_2014941 | 0.08 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr19_-_51014460 | 0.08 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr14_-_64108125 | 0.08 |
ENST00000267522.3
|
WDR89
|
WD repeat domain 89 |
| chr5_-_74807418 | 0.08 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr6_-_36842784 | 0.08 |
ENST00000373699.5
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr9_-_117150303 | 0.08 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr1_-_179834311 | 0.08 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr1_+_168250194 | 0.08 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr20_-_1569351 | 0.08 |
ENST00000563840.1
|
SIRPB1
|
signal-regulatory protein beta 1 |
| chrX_+_133507327 | 0.07 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr5_-_39203093 | 0.07 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr17_+_38375528 | 0.07 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr4_-_185747188 | 0.07 |
ENST00000507295.1
ENST00000504900.1 ENST00000281455.2 ENST00000454703.2 |
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr6_-_146285455 | 0.07 |
ENST00000367505.2
|
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr11_+_36397528 | 0.07 |
ENST00000311599.5
ENST00000378867.3 |
PRR5L
|
proline rich 5 like |
| chr15_+_84115868 | 0.07 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr5_+_133706865 | 0.07 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr8_+_90914073 | 0.07 |
ENST00000297438.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr18_+_56532100 | 0.07 |
ENST00000588456.1
ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr8_-_101719159 | 0.07 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_-_142166796 | 0.07 |
ENST00000392981.2
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr6_+_25279759 | 0.07 |
ENST00000377969.3
|
LRRC16A
|
leucine rich repeat containing 16A |
| chr21_+_35014829 | 0.07 |
ENST00000451686.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr12_+_41221975 | 0.07 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr14_-_53417732 | 0.07 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr4_+_148538517 | 0.07 |
ENST00000296582.3
ENST00000508208.1 |
TMEM184C
|
transmembrane protein 184C |
| chrX_+_17755696 | 0.07 |
ENST00000419185.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr19_+_36393422 | 0.07 |
ENST00000437550.2
|
HCST
|
hematopoietic cell signal transducer |
| chr17_+_1665345 | 0.07 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.5 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.3 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.0 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:1903515 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:1902523 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.0 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.0 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0042424 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.2 | GO:0035766 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0071918 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.0 | 0.1 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.0 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |