Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
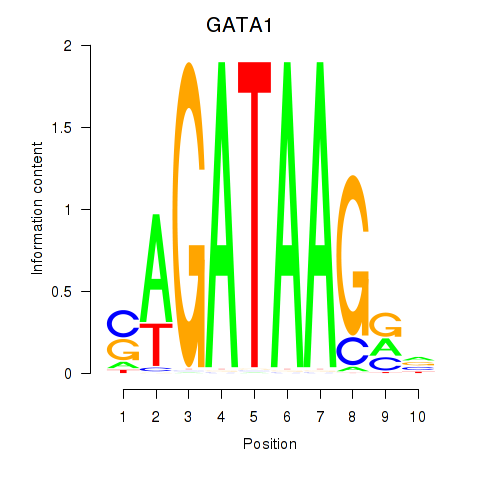
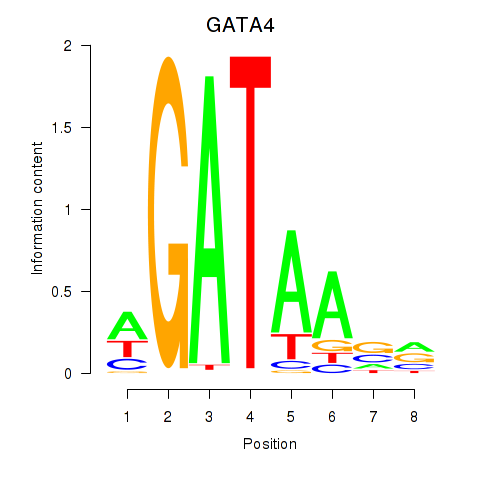
Results for GATA1_GATA4
Z-value: 1.19
Transcription factors associated with GATA1_GATA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA1
|
ENSG00000102145.9 | GATA binding protein 1 |
|
GATA4
|
ENSG00000136574.13 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA1 | hg19_v2_chrX_+_48644962_48644983 | -0.20 | 7.0e-01 | Click! |
Activity profile of GATA1_GATA4 motif
Sorted Z-values of GATA1_GATA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_120502441 | 0.87 |
ENST00000446727.2
|
CCDC64
|
coiled-coil domain containing 64 |
| chr5_+_131409476 | 0.76 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr19_-_13213662 | 0.68 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr4_+_74606223 | 0.67 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr1_-_159880159 | 0.64 |
ENST00000599780.1
|
AL590560.1
|
HCG1995379; Uncharacterized protein |
| chr4_+_165675197 | 0.61 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr3_-_10334585 | 0.58 |
ENST00000430179.1
ENST00000449238.2 ENST00000437422.2 ENST00000287656.7 ENST00000457360.1 ENST00000439975.2 ENST00000446937.2 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr11_-_10920838 | 0.57 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr1_-_235098861 | 0.55 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr6_-_86099898 | 0.54 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr6_+_41010293 | 0.54 |
ENST00000373161.1
ENST00000373158.2 ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chr6_+_155443048 | 0.53 |
ENST00000535583.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_-_107235331 | 0.53 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr8_+_40010989 | 0.50 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr1_-_153013588 | 0.44 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr1_-_109849612 | 0.43 |
ENST00000357155.1
|
MYBPHL
|
myosin binding protein H-like |
| chr1_+_25598872 | 0.41 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr2_+_201994042 | 0.36 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr17_+_70036164 | 0.36 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr12_+_27623565 | 0.34 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr2_-_202298268 | 0.33 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr17_+_38673270 | 0.32 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr3_-_49837254 | 0.32 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr11_-_85393886 | 0.31 |
ENST00000534224.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr10_+_28822417 | 0.31 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr4_-_186732892 | 0.30 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_+_74702214 | 0.30 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr18_-_2982869 | 0.30 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr11_+_64052944 | 0.29 |
ENST00000535675.1
ENST00000543383.1 |
GPR137
|
G protein-coupled receptor 137 |
| chr4_+_76439649 | 0.29 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr1_-_205912577 | 0.29 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr17_+_4853442 | 0.29 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_+_235903480 | 0.29 |
ENST00000454947.1
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr2_+_102928009 | 0.28 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr19_+_37808778 | 0.28 |
ENST00000544914.1
ENST00000589188.1 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr11_+_65222698 | 0.28 |
ENST00000309775.7
|
AP000769.1
|
Uncharacterized protein |
| chr7_-_99381798 | 0.28 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr7_-_100239132 | 0.27 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr14_+_24584056 | 0.27 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr6_+_12290586 | 0.26 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr21_-_37852359 | 0.26 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr17_-_79817091 | 0.25 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr7_+_139528952 | 0.25 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chrX_+_107288197 | 0.25 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_-_116708302 | 0.24 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr11_+_64053005 | 0.24 |
ENST00000538032.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr17_+_19314505 | 0.24 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr19_+_37808831 | 0.24 |
ENST00000589801.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr14_+_24584372 | 0.24 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr11_+_75428857 | 0.24 |
ENST00000198801.5
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2 |
| chr3_+_156807663 | 0.24 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr6_-_138539627 | 0.24 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr17_-_73839792 | 0.24 |
ENST00000590762.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr10_+_71075516 | 0.24 |
ENST00000436817.1
|
HK1
|
hexokinase 1 |
| chr2_+_197504278 | 0.24 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr3_-_10334617 | 0.23 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.8 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chrX_+_107288239 | 0.23 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr8_+_32579321 | 0.23 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr11_-_61734599 | 0.23 |
ENST00000532601.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr5_+_125695805 | 0.22 |
ENST00000513040.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr1_-_219615984 | 0.22 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr3_+_172361483 | 0.22 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr5_-_141703713 | 0.22 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr11_+_64052294 | 0.22 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_+_81106951 | 0.21 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr12_-_67197760 | 0.21 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr7_-_50860565 | 0.21 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
| chr19_+_29704142 | 0.21 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chrX_+_109602039 | 0.21 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr2_+_162165038 | 0.21 |
ENST00000437630.1
|
PSMD14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr5_-_147286065 | 0.21 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr3_+_169629354 | 0.21 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr11_-_64949305 | 0.20 |
ENST00000526623.1
|
AP003068.23
|
Uncharacterized protein |
| chr5_+_36608280 | 0.20 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_+_48516463 | 0.20 |
ENST00000546465.1
|
PFKM
|
phosphofructokinase, muscle |
| chr20_+_51588873 | 0.20 |
ENST00000371497.5
|
TSHZ2
|
teashirt zinc finger homeobox 2 |
| chr17_-_73840614 | 0.20 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr16_+_68071816 | 0.20 |
ENST00000562246.1
|
DUS2
|
dihydrouridine synthase 2 |
| chr1_+_207039154 | 0.20 |
ENST00000367096.3
ENST00000391930.2 |
IL20
|
interleukin 20 |
| chr18_-_8337038 | 0.19 |
ENST00000594251.1
|
AP001094.1
|
Uncharacterized protein |
| chr20_-_44298878 | 0.19 |
ENST00000324384.3
ENST00000356562.2 |
WFDC11
|
WAP four-disulfide core domain 11 |
| chr19_-_51568324 | 0.19 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr9_+_33795533 | 0.19 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chrX_+_100224676 | 0.19 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr17_+_75449889 | 0.19 |
ENST00000590938.1
|
SEPT9
|
septin 9 |
| chr11_+_64052454 | 0.19 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr6_-_107235287 | 0.19 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr11_+_10477733 | 0.19 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr4_-_122148620 | 0.18 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr12_-_120765565 | 0.18 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr11_-_82746587 | 0.18 |
ENST00000528379.1
ENST00000534103.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr4_-_186696561 | 0.17 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_-_91618285 | 0.17 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr14_+_88490894 | 0.17 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr1_-_120354079 | 0.17 |
ENST00000354219.1
ENST00000369401.4 ENST00000256585.5 |
REG4
|
regenerating islet-derived family, member 4 |
| chr21_+_33671160 | 0.17 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr15_+_36887069 | 0.17 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr5_-_20575959 | 0.17 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr1_-_52520828 | 0.17 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr19_-_38344165 | 0.17 |
ENST00000592714.1
ENST00000587395.1 |
AC016582.2
|
AC016582.2 |
| chr20_+_43029911 | 0.17 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr1_-_94050668 | 0.16 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chrX_-_117119243 | 0.16 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr12_-_21928515 | 0.16 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr22_-_30642728 | 0.16 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr17_-_65235916 | 0.16 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr7_-_99381884 | 0.16 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr6_+_33168189 | 0.16 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr6_-_107235378 | 0.16 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr1_-_27240455 | 0.16 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr6_+_31105426 | 0.15 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr11_+_7595136 | 0.15 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr2_+_192543153 | 0.15 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr10_-_4285835 | 0.15 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr9_+_90341024 | 0.15 |
ENST00000340342.6
ENST00000342020.5 |
CTSL
|
cathepsin L |
| chr21_+_39628780 | 0.14 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_+_129720227 | 0.14 |
ENST00000524567.1
|
TMEM45B
|
transmembrane protein 45B |
| chr3_+_49711391 | 0.14 |
ENST00000296456.5
ENST00000449966.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr5_-_33297946 | 0.14 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chr2_+_201994208 | 0.14 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_99833755 | 0.14 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr1_+_66797687 | 0.14 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr11_-_57102947 | 0.14 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr4_-_89619386 | 0.14 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr15_+_59279851 | 0.14 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr6_-_52441713 | 0.14 |
ENST00000182527.3
|
TRAM2
|
translocation associated membrane protein 2 |
| chr8_+_124194875 | 0.14 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr17_-_47045949 | 0.14 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr3_-_139195350 | 0.14 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr8_+_29605860 | 0.14 |
ENST00000517491.1
|
AC145110.1
|
AC145110.1 |
| chr17_-_40288449 | 0.14 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr11_-_1036706 | 0.14 |
ENST00000421673.2
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr16_+_25228242 | 0.14 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr11_-_10920714 | 0.14 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr22_+_35462129 | 0.14 |
ENST00000404699.2
ENST00000308700.6 |
ISX
|
intestine-specific homeobox |
| chr2_-_152830441 | 0.14 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr21_-_36421401 | 0.14 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr4_+_6910966 | 0.14 |
ENST00000444368.1
|
TBC1D14
|
TBC1 domain family, member 14 |
| chr9_-_88896977 | 0.13 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr11_-_105892937 | 0.13 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr1_-_224624730 | 0.13 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr19_-_3772209 | 0.13 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr7_+_8474150 | 0.13 |
ENST00000602349.1
|
NXPH1
|
neurexophilin 1 |
| chr11_+_44748361 | 0.13 |
ENST00000533202.1
ENST00000533080.1 ENST00000520358.2 ENST00000520999.2 |
TSPAN18
|
tetraspanin 18 |
| chr12_+_111843749 | 0.13 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr3_+_42885450 | 0.13 |
ENST00000492609.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr20_-_52645231 | 0.13 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr5_-_135290651 | 0.13 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr15_+_64752927 | 0.13 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chr4_-_144940477 | 0.13 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr8_-_38008783 | 0.13 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chrX_+_118892545 | 0.13 |
ENST00000343905.3
|
SOWAHD
|
sosondowah ankyrin repeat domain family member D |
| chr7_-_2883928 | 0.13 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr1_+_234509413 | 0.13 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr15_-_53002007 | 0.13 |
ENST00000561490.1
|
FAM214A
|
family with sequence similarity 214, member A |
| chr4_-_52883786 | 0.13 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr17_+_33914276 | 0.13 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr10_+_31610064 | 0.13 |
ENST00000446923.2
ENST00000559476.1 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr9_+_71357171 | 0.12 |
ENST00000440050.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr8_-_116504448 | 0.12 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr1_+_161136180 | 0.12 |
ENST00000352210.5
ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX
|
protoporphyrinogen oxidase |
| chr2_+_242289502 | 0.12 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr17_+_41158742 | 0.12 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr19_+_11650709 | 0.12 |
ENST00000586059.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr4_+_79475019 | 0.12 |
ENST00000508214.1
|
ANXA3
|
annexin A3 |
| chr10_+_71075552 | 0.12 |
ENST00000298649.3
|
HK1
|
hexokinase 1 |
| chr6_+_130339710 | 0.12 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr11_+_18154059 | 0.12 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr8_+_70404996 | 0.12 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr6_+_30614886 | 0.12 |
ENST00000376471.4
|
C6orf136
|
chromosome 6 open reading frame 136 |
| chr3_+_185046676 | 0.12 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr17_+_46537697 | 0.12 |
ENST00000579972.1
|
RP11-433M22.2
|
RP11-433M22.2 |
| chr17_-_41050716 | 0.12 |
ENST00000417193.1
ENST00000301683.3 ENST00000436546.1 ENST00000431109.2 |
LINC00671
|
long intergenic non-protein coding RNA 671 |
| chr17_+_75450099 | 0.12 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr8_-_116681686 | 0.12 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr3_-_119379427 | 0.12 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr11_-_66206260 | 0.12 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr7_+_116654935 | 0.12 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chrX_-_43832711 | 0.12 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr1_+_25598989 | 0.12 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chrX_-_19688475 | 0.11 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr13_-_46425865 | 0.11 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr7_+_114055052 | 0.11 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr1_+_145516252 | 0.11 |
ENST00000369306.3
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr17_-_55911970 | 0.11 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr2_+_102927962 | 0.11 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr8_-_128231299 | 0.11 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr1_+_150954493 | 0.11 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr2_-_70520832 | 0.11 |
ENST00000454893.1
ENST00000272348.2 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr8_-_72268721 | 0.11 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chrX_-_78622805 | 0.11 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr6_+_21666633 | 0.11 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr15_+_67420441 | 0.11 |
ENST00000558894.1
|
SMAD3
|
SMAD family member 3 |
| chr10_+_118305435 | 0.11 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr1_+_33283246 | 0.11 |
ENST00000526230.1
ENST00000531256.1 ENST00000482212.1 |
S100PBP
|
S100P binding protein |
| chr2_+_114163945 | 0.11 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr19_-_36822551 | 0.11 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr12_-_56120838 | 0.11 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr3_-_108672742 | 0.11 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr5_+_147443534 | 0.11 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA1_GATA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1904344 | positive regulation of growth rate(GO:0040010) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.3 | 0.8 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.5 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.3 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.2 | GO:0032431 | positive regulation of phospholipase A2 activity(GO:0032430) activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.3 | GO:0060584 | nitric oxide transport(GO:0030185) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.2 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.1 | 0.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 0.7 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.5 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.3 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0018963 | insecticide metabolic process(GO:0017143) phthalate metabolic process(GO:0018963) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.3 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.1 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.1 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.1 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.1 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.0 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.1 | GO:0014740 | regulation of muscle hyperplasia(GO:0014738) negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.0 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.0 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.0 | GO:1903717 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.3 | GO:0043090 | amino acid import(GO:0043090) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.3 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.0 | GO:0036027 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.4 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.3 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.2 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.4 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0034617 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |