Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
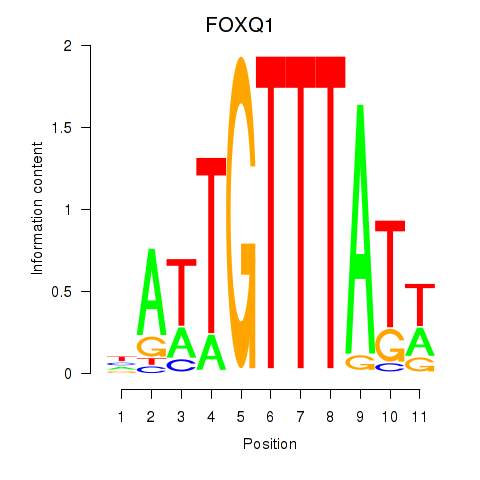
Results for FOXQ1
Z-value: 0.42
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.4 | forkhead box Q1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg19_v2_chr6_+_1312675_1312701 | -0.94 | 4.9e-03 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_41742697 | 0.63 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr1_-_27998689 | 0.59 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr6_+_131958436 | 0.43 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr10_-_4285835 | 0.31 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr2_+_66918558 | 0.29 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr8_+_67104323 | 0.27 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr21_+_30968360 | 0.23 |
ENST00000333765.4
|
GRIK1-AS2
|
GRIK1 antisense RNA 2 |
| chr2_+_152214098 | 0.21 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr5_-_13944652 | 0.21 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr5_+_142286887 | 0.20 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr3_+_98699880 | 0.20 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr22_-_39636914 | 0.20 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr7_+_142985467 | 0.17 |
ENST00000392925.2
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr21_+_42742429 | 0.16 |
ENST00000418103.1
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr9_-_21368075 | 0.16 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr4_-_74853897 | 0.15 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr10_-_21661870 | 0.15 |
ENST00000433460.1
|
RP11-275N1.1
|
RP11-275N1.1 |
| chr10_-_4285923 | 0.15 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr14_-_89878369 | 0.15 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr7_+_28452130 | 0.14 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr4_-_137842536 | 0.14 |
ENST00000512039.1
|
RP11-138I17.1
|
RP11-138I17.1 |
| chr10_+_75668916 | 0.14 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr6_-_133084580 | 0.14 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr15_-_39486510 | 0.14 |
ENST00000560743.1
|
RP11-265N7.1
|
RP11-265N7.1 |
| chr6_-_152623231 | 0.14 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr22_-_39637135 | 0.13 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr9_+_82267508 | 0.13 |
ENST00000490347.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr12_-_49582593 | 0.13 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr4_+_147145709 | 0.13 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr2_+_105050794 | 0.13 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr8_+_94767109 | 0.13 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr16_+_28835766 | 0.12 |
ENST00000564656.1
|
ATXN2L
|
ataxin 2-like |
| chr1_-_151148442 | 0.12 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr2_-_240230890 | 0.12 |
ENST00000446876.1
|
HDAC4
|
histone deacetylase 4 |
| chr2_-_61389168 | 0.12 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr11_-_102668879 | 0.11 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr12_-_49582978 | 0.11 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr11_+_62556596 | 0.11 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr2_+_13677795 | 0.11 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr12_-_102133191 | 0.11 |
ENST00000392924.1
ENST00000266743.2 ENST00000392927.3 |
SYCP3
|
synaptonemal complex protein 3 |
| chr2_+_143635067 | 0.11 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr11_-_615942 | 0.11 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr6_+_31105426 | 0.11 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr17_-_295730 | 0.11 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr11_-_615570 | 0.11 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr20_+_44035847 | 0.11 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_+_173955327 | 0.11 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr2_+_143635222 | 0.11 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr12_-_49582837 | 0.10 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr6_+_4773205 | 0.10 |
ENST00000440139.1
|
CDYL
|
chromodomain protein, Y-like |
| chr4_-_185694872 | 0.10 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr6_+_123038689 | 0.10 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr2_-_203735484 | 0.10 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr20_+_44035200 | 0.10 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_+_10476851 | 0.09 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr4_+_75174204 | 0.09 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chrX_+_107288197 | 0.09 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_-_61389240 | 0.09 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr4_+_165675197 | 0.09 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr6_-_11382478 | 0.09 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr1_+_70820451 | 0.09 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr14_+_52456327 | 0.09 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr10_+_86184676 | 0.09 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr8_-_37411648 | 0.09 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr11_-_102826434 | 0.09 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr21_-_16254231 | 0.09 |
ENST00000412426.1
ENST00000418954.1 |
AF127936.7
|
AF127936.7 |
| chr10_-_94301107 | 0.09 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr21_+_33671264 | 0.08 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr21_+_39628852 | 0.08 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_-_47447970 | 0.08 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr6_+_126221034 | 0.08 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr8_+_94767072 | 0.08 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr7_-_107968999 | 0.08 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr12_+_19928380 | 0.08 |
ENST00000535764.1
|
RP11-405A12.2
|
RP11-405A12.2 |
| chr12_+_58176525 | 0.08 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chrX_+_107288239 | 0.08 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr15_+_57891609 | 0.08 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr12_-_116714564 | 0.08 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr4_-_80994619 | 0.08 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr14_+_77843459 | 0.08 |
ENST00000216471.4
|
SAMD15
|
sterile alpha motif domain containing 15 |
| chr4_-_22444733 | 0.08 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr12_-_92539614 | 0.08 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr3_+_171757346 | 0.08 |
ENST00000421757.1
ENST00000415807.2 ENST00000392699.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr16_-_87729753 | 0.08 |
ENST00000538868.1
|
AC010536.1
|
Uncharacterized protein; cDNA FLJ45526 fis, clone BRTHA2027227 |
| chr11_+_110001723 | 0.08 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr7_+_139528952 | 0.08 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr21_-_16135411 | 0.07 |
ENST00000435315.2
|
AF127936.5
|
AF127936.5 |
| chr1_+_165797024 | 0.07 |
ENST00000372212.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr10_-_101380121 | 0.07 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr21_-_27945562 | 0.07 |
ENST00000299340.4
ENST00000435845.2 |
CYYR1
|
cysteine/tyrosine-rich 1 |
| chr1_+_145524891 | 0.07 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr4_-_21950356 | 0.07 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr5_-_115872124 | 0.07 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr4_+_117220016 | 0.07 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr4_+_153021899 | 0.07 |
ENST00000509332.1
ENST00000504144.1 ENST00000499452.2 |
RP11-18H21.1
|
RP11-18H21.1 |
| chr8_-_74495065 | 0.07 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr13_+_22245522 | 0.07 |
ENST00000382353.5
|
FGF9
|
fibroblast growth factor 9 |
| chr3_-_49066811 | 0.07 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr12_-_47473425 | 0.07 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr4_-_76957214 | 0.07 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr10_+_67330096 | 0.07 |
ENST00000433152.4
ENST00000601979.1 ENST00000599409.1 ENST00000608678.1 |
RP11-222A11.1
|
RP11-222A11.1 |
| chr21_+_33671160 | 0.07 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_+_185431080 | 0.07 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr20_+_56964169 | 0.07 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr1_+_149239529 | 0.07 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr1_+_206972215 | 0.06 |
ENST00000340758.2
|
IL19
|
interleukin 19 |
| chr4_+_17578815 | 0.06 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr17_+_9745786 | 0.06 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr22_-_18257249 | 0.06 |
ENST00000399765.1
ENST00000399767.1 ENST00000399774.3 |
BID
|
BH3 interacting domain death agonist |
| chr6_+_37321823 | 0.06 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr7_+_44646177 | 0.06 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr14_+_35591928 | 0.06 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr14_+_74417192 | 0.06 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr17_-_79212884 | 0.06 |
ENST00000300714.3
|
ENTHD2
|
ENTH domain containing 2 |
| chr11_+_22688150 | 0.06 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr20_-_31989307 | 0.06 |
ENST00000473997.1
ENST00000544843.1 ENST00000346416.2 ENST00000357886.4 ENST00000339269.5 |
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1 |
| chr5_+_73109339 | 0.06 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr4_-_185139062 | 0.06 |
ENST00000296741.2
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr16_+_4784273 | 0.06 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr4_-_68749699 | 0.06 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr6_+_21666633 | 0.06 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr14_+_52456193 | 0.06 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr14_+_74416989 | 0.06 |
ENST00000334571.2
ENST00000554920.1 |
COQ6
|
coenzyme Q6 monooxygenase |
| chr7_+_139529040 | 0.06 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr22_+_24828087 | 0.06 |
ENST00000472248.1
ENST00000436735.1 |
ADORA2A
|
adenosine A2a receptor |
| chr5_-_159766528 | 0.06 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr9_-_127358087 | 0.06 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr10_-_104913367 | 0.06 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr6_-_31648150 | 0.06 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr11_+_108535752 | 0.06 |
ENST00000322536.3
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr9_-_98079965 | 0.06 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr20_+_48429356 | 0.06 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr3_-_155011483 | 0.06 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr11_-_117800080 | 0.06 |
ENST00000524993.1
ENST00000528626.1 ENST00000445164.2 ENST00000430170.2 ENST00000526090.1 |
TMPRSS13
|
transmembrane protease, serine 13 |
| chr1_-_33430286 | 0.06 |
ENST00000373456.7
ENST00000356990.5 ENST00000235150.4 |
RNF19B
|
ring finger protein 19B |
| chr7_+_44646162 | 0.06 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr20_+_36373032 | 0.06 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr19_-_46148820 | 0.06 |
ENST00000587152.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr15_+_89182178 | 0.06 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chrX_+_108780347 | 0.06 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr2_-_165424973 | 0.06 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr5_-_115872142 | 0.05 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chrM_+_10758 | 0.05 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrX_+_10126488 | 0.05 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr10_+_35484793 | 0.05 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr6_+_89674246 | 0.05 |
ENST00000369474.1
|
AL079342.1
|
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr3_-_100565249 | 0.05 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr16_-_69418553 | 0.05 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr12_+_1738363 | 0.05 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr3_-_15382875 | 0.05 |
ENST00000408919.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr20_-_33732952 | 0.05 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr3_+_139063372 | 0.05 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr4_-_19458597 | 0.05 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr3_-_57530051 | 0.05 |
ENST00000311202.6
ENST00000351747.2 ENST00000495027.1 ENST00000389536.4 |
DNAH12
|
dynein, axonemal, heavy chain 12 |
| chr19_+_50433476 | 0.05 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr5_-_139937895 | 0.05 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr14_+_21566980 | 0.05 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr17_-_73851285 | 0.05 |
ENST00000589642.1
ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2
|
WW domain binding protein 2 |
| chr6_-_28973037 | 0.05 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr14_-_39417410 | 0.05 |
ENST00000557019.1
ENST00000556116.1 ENST00000554732.1 |
LINC00639
|
long intergenic non-protein coding RNA 639 |
| chr1_+_153003671 | 0.05 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr10_+_96522361 | 0.05 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr6_-_128841503 | 0.05 |
ENST00000368215.3
ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK
|
protein tyrosine phosphatase, receptor type, K |
| chr17_+_45771420 | 0.05 |
ENST00000578982.1
|
TBKBP1
|
TBK1 binding protein 1 |
| chr1_+_48569306 | 0.05 |
ENST00000601108.1
|
AL109659.1
|
HCG1780467; PRO0529; Uncharacterized protein |
| chr9_+_93759317 | 0.05 |
ENST00000563268.1
|
RP11-367F23.2
|
RP11-367F23.2 |
| chr5_+_138611798 | 0.05 |
ENST00000502394.1
|
MATR3
|
matrin 3 |
| chr2_+_47454054 | 0.05 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr15_+_28624878 | 0.05 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr11_+_34654011 | 0.05 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr4_-_170897045 | 0.05 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr11_+_45943169 | 0.05 |
ENST00000529052.1
ENST00000531526.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr5_-_20575959 | 0.05 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr6_-_131277510 | 0.05 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr5_-_93954227 | 0.05 |
ENST00000513200.3
ENST00000329378.7 ENST00000312498.7 |
KIAA0825
|
KIAA0825 |
| chr2_+_113816685 | 0.05 |
ENST00000393200.2
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr7_+_102988082 | 0.05 |
ENST00000292644.3
ENST00000544811.1 |
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
| chr10_-_11574274 | 0.05 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr17_-_72772462 | 0.05 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr12_-_96389702 | 0.05 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr2_-_4703793 | 0.05 |
ENST00000421212.1
ENST00000412134.1 |
AC022311.1
|
AC022311.1 |
| chr7_-_64023410 | 0.05 |
ENST00000447137.2
|
ZNF680
|
zinc finger protein 680 |
| chr11_+_101983176 | 0.05 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr16_-_25122785 | 0.05 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr12_+_122880045 | 0.05 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr13_+_76378407 | 0.05 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr18_+_66465475 | 0.05 |
ENST00000581520.1
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr4_-_80994210 | 0.05 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr17_+_8339164 | 0.05 |
ENST00000582665.1
ENST00000334527.7 ENST00000299734.7 |
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr16_+_9449445 | 0.05 |
ENST00000564305.1
|
RP11-243A14.1
|
RP11-243A14.1 |
| chr7_-_143582450 | 0.05 |
ENST00000485416.1
|
FAM115A
|
family with sequence similarity 115, member A |
| chr10_+_13652047 | 0.05 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr3_+_173116225 | 0.05 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr7_-_72992865 | 0.05 |
ENST00000452475.1
|
TBL2
|
transducin (beta)-like 2 |
| chr12_+_96588279 | 0.05 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr7_-_7782204 | 0.05 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr17_-_8151353 | 0.05 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr12_+_96588143 | 0.05 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr8_+_103991013 | 0.05 |
ENST00000517983.1
|
KB-1507C5.4
|
KB-1507C5.4 |
| chr3_+_177159695 | 0.05 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.4 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.3 | GO:1990936 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.6 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:1900005 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.0 | GO:1904252 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) negative regulation of bile acid biosynthetic process(GO:0070858) acinar cell differentiation(GO:0090425) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:1903770 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0043474 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.2 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0072544 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.0 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |