Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
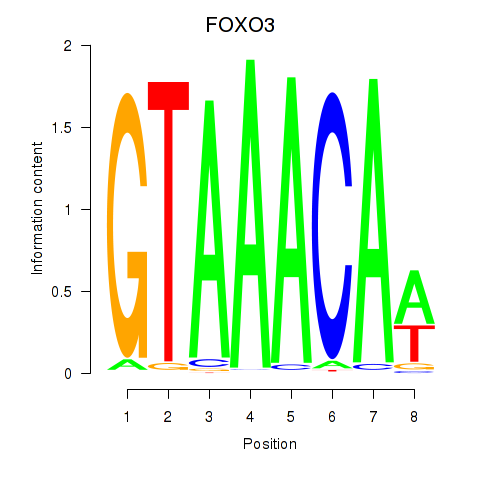
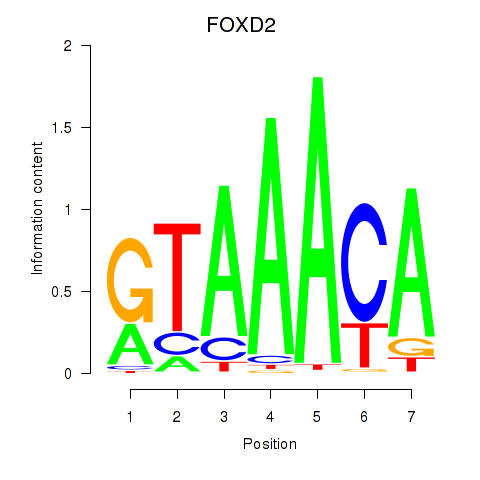
Results for FOXO3_FOXD2
Z-value: 0.68
Transcription factors associated with FOXO3_FOXD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO3
|
ENSG00000118689.10 | forkhead box O3 |
|
FOXD2
|
ENSG00000186564.5 | forkhead box D2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO3 | hg19_v2_chr6_+_108882069_108882087 | 0.42 | 4.1e-01 | Click! |
| FOXD2 | hg19_v2_chr1_+_47901689_47901689 | 0.15 | 7.7e-01 | Click! |
Activity profile of FOXO3_FOXD2 motif
Sorted Z-values of FOXO3_FOXD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_116654376 | 0.81 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr4_+_165675197 | 0.65 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr3_+_178865887 | 0.55 |
ENST00000477735.1
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr6_-_113754604 | 0.55 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr1_+_84630574 | 0.51 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr6_+_21666633 | 0.50 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr3_-_185826718 | 0.43 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr12_+_96588279 | 0.39 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr1_+_120839412 | 0.38 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr14_+_38065052 | 0.37 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr8_+_120079478 | 0.37 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr3_+_172468472 | 0.35 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr3_+_172468505 | 0.29 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr1_+_84629976 | 0.29 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr18_+_61575200 | 0.29 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr3_-_57233966 | 0.29 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr6_+_134758827 | 0.28 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr8_+_71485681 | 0.27 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr5_-_146833803 | 0.27 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr14_-_31926623 | 0.26 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr21_+_39628780 | 0.26 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr6_-_32122106 | 0.26 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr9_-_88896977 | 0.25 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr4_+_165675269 | 0.25 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr12_-_42878101 | 0.25 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr1_+_144989309 | 0.25 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr8_+_95907993 | 0.24 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr5_+_95998714 | 0.23 |
ENST00000506811.1
ENST00000514055.1 |
CAST
|
calpastatin |
| chr7_+_99425633 | 0.23 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr12_-_15038779 | 0.23 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr3_+_69985734 | 0.23 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr7_+_30174668 | 0.23 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_+_198126093 | 0.22 |
ENST00000367385.4
ENST00000442588.1 ENST00000538004.1 |
NEK7
|
NIMA-related kinase 7 |
| chr15_-_31283798 | 0.22 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr18_+_3449330 | 0.22 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr11_+_33061543 | 0.22 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr12_+_64798826 | 0.22 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr4_-_152149033 | 0.21 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr17_+_57408994 | 0.21 |
ENST00000312655.4
|
YPEL2
|
yippee-like 2 (Drosophila) |
| chr14_-_36983034 | 0.21 |
ENST00000518529.2
|
SFTA3
|
surfactant associated 3 |
| chr7_+_77469439 | 0.21 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr3_+_125985620 | 0.21 |
ENST00000511512.1
ENST00000512435.1 |
RP11-71E19.1
|
RP11-71E19.1 |
| chr3_+_187930491 | 0.21 |
ENST00000443217.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_134326009 | 0.21 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr1_+_198126209 | 0.20 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr15_-_99057551 | 0.20 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr12_-_15865844 | 0.20 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr8_-_131028782 | 0.20 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr8_+_26150628 | 0.20 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr9_+_4792971 | 0.20 |
ENST00000381732.3
ENST00000442869.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr8_-_117886563 | 0.20 |
ENST00000519837.1
ENST00000522699.1 |
RAD21
|
RAD21 homolog (S. pombe) |
| chr9_-_3469181 | 0.20 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr5_+_131746575 | 0.19 |
ENST00000337752.2
ENST00000378947.3 ENST00000407797.1 |
C5orf56
|
chromosome 5 open reading frame 56 |
| chr8_-_80993010 | 0.19 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr12_-_102591604 | 0.19 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr2_+_197577841 | 0.19 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr3_+_48956249 | 0.19 |
ENST00000452882.1
ENST00000430423.1 ENST00000356401.4 ENST00000449376.1 ENST00000420814.1 ENST00000449729.1 ENST00000433170.1 |
ARIH2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
| chr11_+_13299186 | 0.19 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr1_+_84630053 | 0.19 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr7_+_28448995 | 0.19 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr4_-_141075330 | 0.19 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr6_+_139094657 | 0.19 |
ENST00000332797.6
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr1_+_20512568 | 0.18 |
ENST00000375099.3
|
UBXN10
|
UBX domain protein 10 |
| chr9_-_135819987 | 0.18 |
ENST00000298552.3
ENST00000403810.1 |
TSC1
|
tuberous sclerosis 1 |
| chr3_-_129375556 | 0.18 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr10_-_14050522 | 0.18 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr3_-_168865522 | 0.18 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr11_-_119247004 | 0.18 |
ENST00000531070.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr12_-_48213735 | 0.18 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr8_-_117886732 | 0.17 |
ENST00000517485.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr20_+_62887139 | 0.17 |
ENST00000609764.1
|
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr5_-_126409159 | 0.17 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr6_-_134639042 | 0.17 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_52869205 | 0.17 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr18_+_9475585 | 0.17 |
ENST00000585015.1
|
RALBP1
|
ralA binding protein 1 |
| chr1_+_84630352 | 0.17 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_+_97759848 | 0.17 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr8_+_97773202 | 0.16 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr17_+_65374075 | 0.16 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr21_-_16135411 | 0.16 |
ENST00000435315.2
|
AF127936.5
|
AF127936.5 |
| chr3_+_159570722 | 0.16 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr7_-_84121858 | 0.16 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr12_+_13349650 | 0.16 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr10_-_4720333 | 0.15 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr14_+_24584372 | 0.15 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr19_-_18902106 | 0.15 |
ENST00000542601.2
ENST00000425807.1 ENST00000222271.2 |
COMP
|
cartilage oligomeric matrix protein |
| chr18_+_66465302 | 0.15 |
ENST00000360242.5
ENST00000358653.5 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr20_+_44035847 | 0.15 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr13_+_50589390 | 0.15 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr17_-_26662464 | 0.15 |
ENST00000579419.1
ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr11_-_71752571 | 0.15 |
ENST00000544238.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr1_-_179851611 | 0.15 |
ENST00000610272.1
|
RP11-533E19.7
|
RP11-533E19.7 |
| chr3_+_178866199 | 0.14 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr14_-_65569057 | 0.14 |
ENST00000555419.1
ENST00000341653.2 |
MAX
|
MYC associated factor X |
| chr10_-_49482907 | 0.14 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr6_-_13621126 | 0.14 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr11_-_65150103 | 0.14 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr2_+_66918558 | 0.14 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr14_-_58894332 | 0.14 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr3_-_46608010 | 0.14 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr8_-_117886612 | 0.14 |
ENST00000520992.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr17_+_72733350 | 0.14 |
ENST00000392613.5
ENST00000392612.3 ENST00000392610.1 |
RAB37
|
RAB37, member RAS oncogene family |
| chr13_+_52598827 | 0.14 |
ENST00000521776.2
|
UTP14C
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr20_-_45530365 | 0.14 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr2_+_173955327 | 0.14 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr16_+_21623958 | 0.14 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr5_+_95998746 | 0.14 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr11_+_10476851 | 0.14 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr5_+_139739772 | 0.14 |
ENST00000506757.2
ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr17_-_30185946 | 0.14 |
ENST00000579741.1
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr14_-_102605983 | 0.13 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr7_-_140624499 | 0.13 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr4_-_89442940 | 0.13 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr3_-_185826855 | 0.13 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr3_-_33700933 | 0.13 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chrX_+_78003204 | 0.13 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr10_+_99079008 | 0.13 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr6_-_56716686 | 0.13 |
ENST00000520645.1
|
DST
|
dystonin |
| chr11_+_62186498 | 0.13 |
ENST00000278282.2
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr2_+_111878483 | 0.13 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr11_-_71752838 | 0.13 |
ENST00000537930.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr6_+_74405501 | 0.13 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr10_+_95848824 | 0.13 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr17_+_57642886 | 0.13 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr2_+_28974489 | 0.13 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr6_+_151358048 | 0.13 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr9_+_132044730 | 0.13 |
ENST00000455981.1
|
RP11-344B5.2
|
RP11-344B5.2 |
| chr6_+_32121789 | 0.12 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr9_-_3525968 | 0.12 |
ENST00000382004.3
ENST00000302303.1 ENST00000449190.1 |
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr7_+_116654935 | 0.12 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr4_-_87279641 | 0.12 |
ENST00000512689.1
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_+_109223595 | 0.12 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr16_-_69418649 | 0.12 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr8_-_117886955 | 0.12 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr6_+_74405804 | 0.12 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr17_+_70026795 | 0.12 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr14_-_58893832 | 0.12 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr7_+_30174426 | 0.12 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr11_-_85780853 | 0.12 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_+_90339169 | 0.12 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr7_+_129906660 | 0.12 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr14_-_65569186 | 0.12 |
ENST00000555932.1
ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX
|
MYC associated factor X |
| chr14_-_73493784 | 0.12 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr16_+_2880254 | 0.12 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr8_-_116673894 | 0.12 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr10_+_23728198 | 0.12 |
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1 |
| chr1_+_84630367 | 0.12 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_-_179112189 | 0.12 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr12_+_25348186 | 0.12 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr8_-_99954788 | 0.12 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr15_-_31283618 | 0.11 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr6_-_113953705 | 0.11 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr1_+_206138884 | 0.11 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chr17_+_65373531 | 0.11 |
ENST00000580974.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr10_-_98347063 | 0.11 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr19_-_6502304 | 0.11 |
ENST00000540257.1
ENST00000594276.1 ENST00000594075.1 ENST00000600216.1 ENST00000596926.1 |
TUBB4A
|
tubulin, beta 4A class IVa |
| chr1_+_3541543 | 0.11 |
ENST00000378344.2
ENST00000344579.5 |
TPRG1L
|
tumor protein p63 regulated 1-like |
| chr10_-_32217717 | 0.11 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr1_+_24646263 | 0.11 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr3_+_69928256 | 0.11 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_200335983 | 0.11 |
ENST00000457245.1
|
SATB2
|
SATB homeobox 2 |
| chr15_-_33360342 | 0.11 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr17_+_67410832 | 0.11 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_-_45956868 | 0.11 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr10_-_4720301 | 0.11 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr16_+_86612112 | 0.11 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr13_-_31038370 | 0.11 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr10_-_45474237 | 0.11 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr18_-_59415987 | 0.11 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr2_-_71454185 | 0.11 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr1_+_110158726 | 0.11 |
ENST00000531734.1
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr2_-_198175495 | 0.10 |
ENST00000409153.1
ENST00000409919.1 ENST00000539527.1 |
ANKRD44
|
ankyrin repeat domain 44 |
| chr17_-_46035187 | 0.10 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr14_+_56127989 | 0.10 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr14_+_24584056 | 0.10 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr6_+_32121908 | 0.10 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr8_-_29592736 | 0.10 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr4_-_186732892 | 0.10 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_+_74903580 | 0.10 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr5_+_95998673 | 0.10 |
ENST00000514845.1
|
CAST
|
calpastatin |
| chr12_+_13349711 | 0.10 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr6_+_116850174 | 0.10 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr10_-_46089939 | 0.10 |
ENST00000453980.3
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr6_+_144980954 | 0.10 |
ENST00000367525.3
|
UTRN
|
utrophin |
| chrX_-_20237059 | 0.10 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr14_+_32798547 | 0.10 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr3_+_9691117 | 0.10 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr14_-_65569244 | 0.10 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr14_+_56127960 | 0.10 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr15_-_70994612 | 0.10 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr5_-_59064458 | 0.10 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_+_72251857 | 0.10 |
ENST00000507345.2
ENST00000512348.1 ENST00000287761.6 |
FCHO2
|
FCH domain only 2 |
| chr1_+_209878182 | 0.10 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr14_-_75518129 | 0.10 |
ENST00000556257.1
ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3
|
mutL homolog 3 |
| chr15_+_36994210 | 0.10 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr22_-_37880543 | 0.10 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_84630645 | 0.10 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_-_61777090 | 0.10 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr12_+_79258444 | 0.10 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr17_+_9745786 | 0.10 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO3_FOXD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 1.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.6 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.6 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.2 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:1902868 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.0 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.0 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.3 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.4 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:1903823 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.0 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.0 | 0.3 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.0 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.0 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0072501 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.0 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |