Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
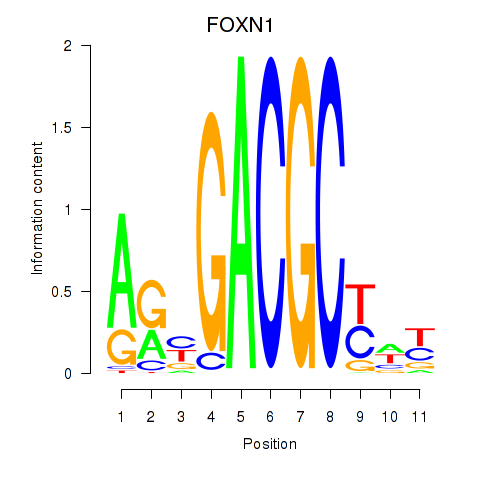
Results for FOXN1
Z-value: 0.86
Transcription factors associated with FOXN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXN1
|
ENSG00000109101.3 | forkhead box N1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXN1 | hg19_v2_chr17_+_26833250_26833278 | 0.28 | 5.9e-01 | Click! |
Activity profile of FOXN1 motif
Sorted Z-values of FOXN1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_54198210 | 0.45 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chrX_-_108868390 | 0.43 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr14_-_77495007 | 0.43 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr2_+_46769798 | 0.42 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr2_-_38830030 | 0.38 |
ENST00000410076.1
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chrX_-_83442915 | 0.37 |
ENST00000262752.2
ENST00000543399.1 |
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr19_-_56110859 | 0.34 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chrX_-_106959631 | 0.32 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr8_+_38243721 | 0.30 |
ENST00000527334.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chrX_+_105969893 | 0.28 |
ENST00000255499.2
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr15_+_71184931 | 0.27 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chrX_+_153237740 | 0.27 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chr1_+_74663896 | 0.27 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chr1_-_245134273 | 0.26 |
ENST00000607453.1
|
RP11-156E8.1
|
Uncharacterized protein |
| chr11_-_82782952 | 0.26 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr20_-_36156264 | 0.26 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr18_+_9136758 | 0.25 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr13_-_95364389 | 0.25 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr10_+_14880287 | 0.25 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr17_-_56065540 | 0.25 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr17_-_58469591 | 0.24 |
ENST00000589335.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chrX_-_117107680 | 0.24 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr3_+_52280220 | 0.24 |
ENST00000409502.3
ENST00000323588.4 |
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr11_-_82782861 | 0.24 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr11_+_1860200 | 0.24 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr5_+_149340282 | 0.23 |
ENST00000286298.4
|
SLC26A2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr12_+_56110374 | 0.23 |
ENST00000549424.1
|
RP11-644F5.10
|
Uncharacterized protein |
| chr12_+_88536067 | 0.22 |
ENST00000549011.1
ENST00000266712.6 ENST00000551088.1 |
TMTC3
|
transmembrane and tetratricopeptide repeat containing 3 |
| chr3_-_15469045 | 0.21 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr6_+_27215471 | 0.20 |
ENST00000421826.2
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr6_+_27215494 | 0.20 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr7_-_120498357 | 0.20 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr3_-_27410847 | 0.19 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr2_-_28113217 | 0.19 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr1_-_28241024 | 0.19 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr1_+_101361626 | 0.19 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr5_+_176873789 | 0.19 |
ENST00000323249.3
ENST00000502922.1 |
PRR7
|
proline rich 7 (synaptic) |
| chr5_+_61602055 | 0.19 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr22_+_29469100 | 0.19 |
ENST00000327813.5
ENST00000407188.1 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr14_+_37126765 | 0.18 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr1_-_59012365 | 0.18 |
ENST00000456980.1
ENST00000482274.2 ENST00000453710.1 ENST00000419242.1 ENST00000358603.2 ENST00000371226.3 ENST00000426139.1 |
OMA1
|
OMA1 zinc metallopeptidase |
| chr2_+_30670077 | 0.17 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr12_-_88535747 | 0.17 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chr12_-_52967600 | 0.17 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr19_-_37663572 | 0.17 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chr1_+_206138457 | 0.16 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr3_+_180319918 | 0.16 |
ENST00000296015.4
ENST00000491380.1 ENST00000412756.2 ENST00000382584.4 |
TTC14
|
tetratricopeptide repeat domain 14 |
| chr11_-_65325430 | 0.16 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr2_+_130939235 | 0.16 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr4_+_164415855 | 0.15 |
ENST00000508268.1
|
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr5_+_61601965 | 0.15 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr14_+_100204027 | 0.15 |
ENST00000554479.1
ENST00000555145.1 |
EML1
|
echinoderm microtubule associated protein like 1 |
| chr3_+_52279902 | 0.15 |
ENST00000457454.1
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr8_+_38243967 | 0.15 |
ENST00000524874.1
ENST00000379957.4 ENST00000523983.2 |
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr1_-_8939265 | 0.15 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr5_+_131892603 | 0.15 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr15_+_71185148 | 0.15 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr4_+_148538517 | 0.15 |
ENST00000296582.3
ENST00000508208.1 |
TMEM184C
|
transmembrane protein 184C |
| chrX_-_117107542 | 0.15 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr19_+_39897943 | 0.14 |
ENST00000600033.1
|
ZFP36
|
ZFP36 ring finger protein |
| chr11_-_73882249 | 0.14 |
ENST00000535954.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr12_+_56110315 | 0.14 |
ENST00000548556.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr3_-_195163584 | 0.14 |
ENST00000439666.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr12_-_88535842 | 0.14 |
ENST00000550962.1
ENST00000552810.1 |
CEP290
|
centrosomal protein 290kDa |
| chr10_+_94352956 | 0.14 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr5_+_61602236 | 0.14 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr19_-_11639910 | 0.14 |
ENST00000588998.1
ENST00000586149.1 |
ECSIT
|
ECSIT signalling integrator |
| chr2_+_30670209 | 0.14 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr10_-_14880002 | 0.14 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr14_-_92588246 | 0.13 |
ENST00000329559.3
|
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr15_-_83735889 | 0.13 |
ENST00000379403.2
|
BTBD1
|
BTB (POZ) domain containing 1 |
| chr15_-_83736091 | 0.13 |
ENST00000261721.4
|
BTBD1
|
BTB (POZ) domain containing 1 |
| chr2_-_68694390 | 0.13 |
ENST00000377957.3
|
FBXO48
|
F-box protein 48 |
| chr17_-_43510282 | 0.13 |
ENST00000290470.3
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr10_+_93558069 | 0.13 |
ENST00000371627.4
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr2_+_190306159 | 0.13 |
ENST00000314761.4
|
WDR75
|
WD repeat domain 75 |
| chr7_-_130353553 | 0.13 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr5_+_93954039 | 0.13 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr4_+_90032651 | 0.13 |
ENST00000603357.1
|
RP11-84C13.1
|
RP11-84C13.1 |
| chr11_-_27384737 | 0.13 |
ENST00000317945.6
|
CCDC34
|
coiled-coil domain containing 34 |
| chr2_-_165697717 | 0.13 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr7_+_100464760 | 0.13 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr1_+_40627038 | 0.13 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr5_+_140019004 | 0.13 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr19_+_33463210 | 0.13 |
ENST00000590281.1
|
C19orf40
|
chromosome 19 open reading frame 40 |
| chr4_-_170533723 | 0.13 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr6_+_7590413 | 0.12 |
ENST00000342415.5
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr6_+_52930237 | 0.12 |
ENST00000323557.7
|
FBXO9
|
F-box protein 9 |
| chr19_-_46389359 | 0.12 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr13_+_73356197 | 0.12 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr4_+_119199864 | 0.12 |
ENST00000602414.1
ENST00000602520.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr19_+_57742369 | 0.12 |
ENST00000415300.2
ENST00000448930.1 |
AURKC
|
aurora kinase C |
| chr11_+_117014983 | 0.12 |
ENST00000527958.1
ENST00000419197.2 ENST00000304808.6 ENST00000529887.2 |
PAFAH1B2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) |
| chr19_-_42806723 | 0.12 |
ENST00000262890.3
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr15_-_52472078 | 0.12 |
ENST00000396335.4
ENST00000560116.1 ENST00000358784.7 |
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr2_+_113403434 | 0.12 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr1_+_221054584 | 0.12 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr6_-_150039249 | 0.12 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr2_-_61765315 | 0.11 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr4_-_1713977 | 0.11 |
ENST00000318386.4
|
SLBP
|
stem-loop binding protein |
| chr17_+_29421987 | 0.11 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr6_-_150039170 | 0.11 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr20_-_57607347 | 0.11 |
ENST00000395663.1
ENST00000395659.1 ENST00000243997.3 |
ATP5E
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chrX_-_99891796 | 0.11 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr20_+_3801162 | 0.11 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr5_+_32710736 | 0.11 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr22_+_29469012 | 0.11 |
ENST00000400335.4
ENST00000400338.2 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr15_-_77712477 | 0.11 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr14_+_65016620 | 0.11 |
ENST00000298705.1
|
PPP1R36
|
protein phosphatase 1, regulatory subunit 36 |
| chr2_+_73612858 | 0.11 |
ENST00000409009.1
ENST00000264448.6 ENST00000377715.1 |
ALMS1
|
Alstrom syndrome 1 |
| chr5_+_80256453 | 0.11 |
ENST00000265080.4
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2 |
| chr10_+_127585093 | 0.11 |
ENST00000368695.1
ENST00000368693.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr12_-_6602955 | 0.11 |
ENST00000543703.1
|
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr2_+_241508039 | 0.11 |
ENST00000270357.4
|
RNPEPL1
|
arginyl aminopeptidase (aminopeptidase B)-like 1 |
| chr12_+_70760056 | 0.11 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr7_+_131012605 | 0.10 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr1_-_74663825 | 0.10 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr3_+_52280173 | 0.10 |
ENST00000296487.4
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr5_+_140019079 | 0.10 |
ENST00000252100.6
|
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr2_-_152684977 | 0.10 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr19_+_12902289 | 0.10 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr11_-_73882029 | 0.10 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr12_+_56110247 | 0.10 |
ENST00000551926.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr12_+_51632638 | 0.10 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr14_+_54863682 | 0.10 |
ENST00000543789.2
ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr22_-_31741757 | 0.10 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr1_+_82266053 | 0.10 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr3_+_9932238 | 0.10 |
ENST00000307768.4
|
JAGN1
|
jagunal homolog 1 (Drosophila) |
| chr17_-_43045439 | 0.10 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr6_-_111136299 | 0.10 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr12_+_56109926 | 0.10 |
ENST00000547076.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr10_-_119134918 | 0.09 |
ENST00000334464.5
|
PDZD8
|
PDZ domain containing 8 |
| chr2_+_170655789 | 0.09 |
ENST00000409333.1
|
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr19_-_48867171 | 0.09 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr13_-_21635631 | 0.09 |
ENST00000382592.4
|
LATS2
|
large tumor suppressor kinase 2 |
| chr14_-_96830207 | 0.09 |
ENST00000359933.4
|
ATG2B
|
autophagy related 2B |
| chr18_-_19180681 | 0.09 |
ENST00000269214.5
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr3_-_49158218 | 0.09 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr2_-_165697920 | 0.09 |
ENST00000342193.4
ENST00000375458.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr12_+_3600356 | 0.09 |
ENST00000382622.3
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr5_-_93447333 | 0.09 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr17_-_58469329 | 0.09 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr9_+_102668915 | 0.09 |
ENST00000259400.6
ENST00000531035.1 ENST00000525640.1 ENST00000534052.1 ENST00000526607.1 |
STX17
|
syntaxin 17 |
| chr17_-_33905521 | 0.09 |
ENST00000225873.4
|
PEX12
|
peroxisomal biogenesis factor 12 |
| chr4_-_103748271 | 0.08 |
ENST00000343106.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_156393349 | 0.08 |
ENST00000473702.1
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr4_+_177241094 | 0.08 |
ENST00000503362.1
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr16_+_30772913 | 0.08 |
ENST00000563909.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr2_-_38830090 | 0.08 |
ENST00000449105.3
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr16_-_30538079 | 0.08 |
ENST00000562803.1
|
ZNF768
|
zinc finger protein 768 |
| chr17_+_29421900 | 0.08 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr17_+_36584662 | 0.08 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr8_+_38243951 | 0.08 |
ENST00000297720.5
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr3_-_15469006 | 0.08 |
ENST00000443029.1
ENST00000383790.3 ENST00000383789.5 |
METTL6
|
methyltransferase like 6 |
| chr2_+_187350973 | 0.08 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr3_+_137906353 | 0.08 |
ENST00000461822.1
ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr3_-_120461353 | 0.08 |
ENST00000483733.1
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr10_+_21823243 | 0.08 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr17_-_64187973 | 0.08 |
ENST00000583358.1
ENST00000392769.2 |
CEP112
|
centrosomal protein 112kDa |
| chr4_+_164415594 | 0.08 |
ENST00000509657.1
ENST00000358572.5 |
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chrX_+_38660682 | 0.08 |
ENST00000457894.1
|
MID1IP1
|
MID1 interacting protein 1 |
| chr3_+_50654821 | 0.08 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr5_+_32585605 | 0.08 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr15_+_59063478 | 0.08 |
ENST00000559228.1
ENST00000450403.2 |
FAM63B
|
family with sequence similarity 63, member B |
| chr16_+_69221028 | 0.08 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr16_-_56458896 | 0.07 |
ENST00000565445.1
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chr1_+_171283331 | 0.07 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr13_-_73356234 | 0.07 |
ENST00000545453.1
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr19_+_42772659 | 0.07 |
ENST00000572681.2
|
CIC
|
capicua transcriptional repressor |
| chr9_+_114393634 | 0.07 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr4_-_10023095 | 0.07 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr3_-_133380731 | 0.07 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr5_-_150138061 | 0.07 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr4_-_1714037 | 0.07 |
ENST00000488267.1
ENST00000429429.2 ENST00000480936.1 |
SLBP
|
stem-loop binding protein |
| chr17_+_40913264 | 0.07 |
ENST00000587142.1
ENST00000588576.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr6_+_166945369 | 0.07 |
ENST00000598601.1
|
Z98049.1
|
CDNA FLJ25492 fis, clone CBR01389; Uncharacterized protein |
| chr13_-_24463530 | 0.07 |
ENST00000382172.3
|
MIPEP
|
mitochondrial intermediate peptidase |
| chr19_-_58662139 | 0.07 |
ENST00000598312.1
|
ZNF329
|
zinc finger protein 329 |
| chr1_+_172502244 | 0.07 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chrX_+_38660704 | 0.07 |
ENST00000378474.3
|
MID1IP1
|
MID1 interacting protein 1 |
| chr19_+_11039391 | 0.07 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr12_+_12938541 | 0.07 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr4_+_119200215 | 0.07 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr20_+_825275 | 0.07 |
ENST00000541082.1
|
FAM110A
|
family with sequence similarity 110, member A |
| chr11_-_133826852 | 0.07 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr10_-_127408011 | 0.07 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chr7_-_148725544 | 0.07 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr13_-_73356009 | 0.07 |
ENST00000377780.4
ENST00000377767.4 |
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr2_+_187350883 | 0.07 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr2_+_10560147 | 0.07 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin-like 1 |
| chr12_+_49740700 | 0.07 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr10_+_5726764 | 0.06 |
ENST00000328090.5
ENST00000496681.1 |
FAM208B
|
family with sequence similarity 208, member B |
| chr1_-_78149041 | 0.06 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr19_+_19144384 | 0.06 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr10_+_127408110 | 0.06 |
ENST00000356792.4
|
C10orf137
|
erythroid differentiation regulatory factor 1 |
| chr1_+_235490659 | 0.06 |
ENST00000488594.1
|
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr7_-_27153454 | 0.06 |
ENST00000522456.1
|
HOXA3
|
homeobox A3 |
| chr10_+_103113840 | 0.06 |
ENST00000393441.4
ENST00000408038.2 |
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr12_+_51632508 | 0.06 |
ENST00000449723.3
|
DAZAP2
|
DAZ associated protein 2 |
| chr3_-_195163803 | 0.06 |
ENST00000326793.6
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXN1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.5 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.0 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.4 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |