Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
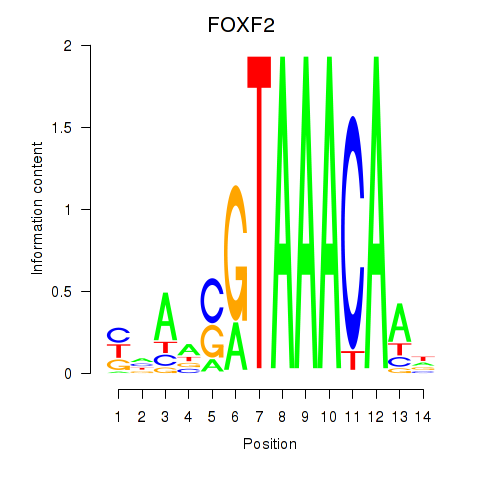
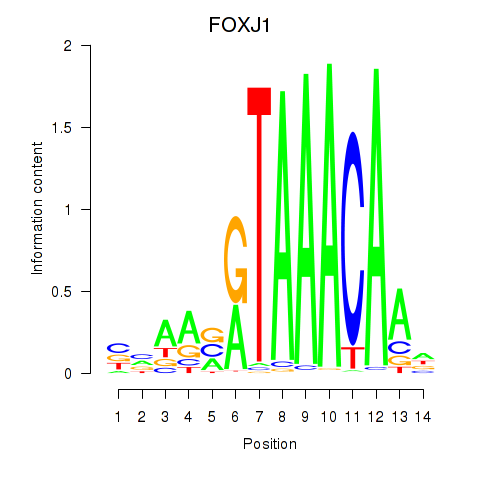
Results for FOXF2_FOXJ1
Z-value: 0.28
Transcription factors associated with FOXF2_FOXJ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXF2
|
ENSG00000137273.3 | forkhead box F2 |
|
FOXJ1
|
ENSG00000129654.7 | forkhead box J1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXF2 | hg19_v2_chr6_+_1389989_1390069 | 0.38 | 4.6e-01 | Click! |
| FOXJ1 | hg19_v2_chr17_-_74137374_74137385 | -0.11 | 8.3e-01 | Click! |
Activity profile of FOXF2_FOXJ1 motif
Sorted Z-values of FOXF2_FOXJ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_77233294 | 0.18 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr5_-_42811986 | 0.16 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr17_+_72427477 | 0.13 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr15_-_76352069 | 0.12 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr17_+_72426891 | 0.10 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr9_+_136287444 | 0.10 |
ENST00000355699.2
ENST00000356589.2 ENST00000371911.3 |
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr11_+_113930955 | 0.10 |
ENST00000535700.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr7_-_105332084 | 0.09 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr5_-_42812143 | 0.09 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr8_-_116673894 | 0.09 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_+_101846664 | 0.09 |
ENST00000421544.1
ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr11_-_119247004 | 0.08 |
ENST00000531070.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr19_+_18492973 | 0.08 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr14_-_36988882 | 0.07 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr8_+_97597148 | 0.07 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr12_-_62585203 | 0.07 |
ENST00000551449.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr17_-_8263538 | 0.07 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr8_+_31496809 | 0.06 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr17_+_67410832 | 0.06 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr7_+_129906660 | 0.06 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr1_-_47131521 | 0.06 |
ENST00000542495.1
ENST00000532925.1 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr5_-_142065612 | 0.06 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chrX_+_9880412 | 0.06 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr7_+_12250886 | 0.06 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr12_+_40787194 | 0.06 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr11_+_844067 | 0.06 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr1_-_57431679 | 0.06 |
ENST00000371237.4
ENST00000535057.1 ENST00000543257.1 |
C8B
|
complement component 8, beta polypeptide |
| chr13_-_41240717 | 0.06 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr15_+_63188009 | 0.06 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr13_-_40924439 | 0.05 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr16_+_777739 | 0.05 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr20_-_22566089 | 0.05 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr9_+_131904233 | 0.05 |
ENST00000432651.1
ENST00000435132.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr19_+_7580103 | 0.05 |
ENST00000596712.1
|
ZNF358
|
zinc finger protein 358 |
| chr11_+_844406 | 0.05 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr15_-_34610962 | 0.05 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr1_+_40862501 | 0.05 |
ENST00000539317.1
|
SMAP2
|
small ArfGAP2 |
| chr5_+_60933634 | 0.05 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr17_-_6947225 | 0.05 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr15_-_34630234 | 0.05 |
ENST00000558667.1
ENST00000561120.1 ENST00000559236.1 ENST00000397702.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr8_+_99956662 | 0.05 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr14_-_31926623 | 0.05 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chrX_+_9880590 | 0.05 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr7_-_112579673 | 0.05 |
ENST00000432572.1
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr15_-_34629922 | 0.04 |
ENST00000559484.1
ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr5_+_137673200 | 0.04 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr9_+_131903916 | 0.04 |
ENST00000419582.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr4_+_71108300 | 0.04 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr2_-_233877912 | 0.04 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr16_-_80926457 | 0.04 |
ENST00000563626.1
ENST00000562231.1 |
RP11-314O13.1
|
RP11-314O13.1 |
| chr2_+_232575168 | 0.04 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr10_-_375422 | 0.04 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr1_+_43735678 | 0.04 |
ENST00000432792.2
|
TMEM125
|
transmembrane protein 125 |
| chr8_-_131028869 | 0.04 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr4_-_140223614 | 0.04 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr4_-_174256276 | 0.04 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr9_+_131904295 | 0.04 |
ENST00000434095.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr7_+_12250943 | 0.04 |
ENST00000442107.1
|
TMEM106B
|
transmembrane protein 106B |
| chr7_-_84122033 | 0.04 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr11_+_33061336 | 0.04 |
ENST00000602733.1
|
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr5_-_41794663 | 0.04 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr11_+_27076764 | 0.04 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_+_43735646 | 0.04 |
ENST00000439858.1
|
TMEM125
|
transmembrane protein 125 |
| chr9_-_134585221 | 0.04 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr6_-_28973037 | 0.04 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr19_+_18496957 | 0.04 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr1_-_1356719 | 0.04 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr17_-_7082668 | 0.03 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr22_-_39928823 | 0.03 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr1_+_149239529 | 0.03 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr1_+_114472992 | 0.03 |
ENST00000514621.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr14_+_74551650 | 0.03 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr18_-_53068940 | 0.03 |
ENST00000562638.1
|
TCF4
|
transcription factor 4 |
| chr7_+_77469439 | 0.03 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr8_+_79428539 | 0.03 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr11_-_8739383 | 0.03 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr13_-_46756351 | 0.03 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr16_+_777246 | 0.03 |
ENST00000561546.1
ENST00000564545.1 ENST00000389703.3 ENST00000567414.1 ENST00000568141.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr19_-_36523529 | 0.03 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr4_+_104346194 | 0.03 |
ENST00000510200.1
|
RP11-328K4.1
|
RP11-328K4.1 |
| chr3_-_107941209 | 0.03 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr12_+_25205666 | 0.03 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr2_-_128615681 | 0.03 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr12_-_63328817 | 0.03 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr2_+_175260514 | 0.03 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr11_-_63933504 | 0.03 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr2_-_192711968 | 0.03 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr1_+_114473350 | 0.03 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr14_-_69262916 | 0.03 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr15_+_42697065 | 0.03 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr12_+_72080253 | 0.03 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr18_-_25616519 | 0.03 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr10_+_70847852 | 0.03 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr1_-_152297679 | 0.03 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr7_+_5229819 | 0.03 |
ENST00000288828.4
ENST00000401525.3 ENST00000404704.3 |
WIPI2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr2_-_207078086 | 0.03 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr4_+_79567314 | 0.03 |
ENST00000503539.1
ENST00000504675.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr7_+_116593292 | 0.03 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr3_+_119298523 | 0.03 |
ENST00000357003.3
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr7_+_134832808 | 0.03 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr7_+_5229904 | 0.03 |
ENST00000382384.2
|
WIPI2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr15_-_31283618 | 0.03 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr2_+_33661382 | 0.03 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_+_176853702 | 0.03 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr3_+_157828152 | 0.03 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr12_-_15374343 | 0.03 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr1_+_92545862 | 0.03 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr1_+_172745006 | 0.02 |
ENST00000432694.2
|
RP1-15D23.2
|
RP1-15D23.2 |
| chr11_-_16419067 | 0.02 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_14541060 | 0.02 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr3_+_159570722 | 0.02 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr22_-_37505449 | 0.02 |
ENST00000406725.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chr14_-_74551096 | 0.02 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr14_+_32964258 | 0.02 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_25664408 | 0.02 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr18_-_53068782 | 0.02 |
ENST00000569012.1
|
TCF4
|
transcription factor 4 |
| chr5_-_137674000 | 0.02 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr20_-_25320367 | 0.02 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr1_+_15668240 | 0.02 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr4_-_70626314 | 0.02 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr15_-_31283798 | 0.02 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr2_-_207078154 | 0.02 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr6_-_110964453 | 0.02 |
ENST00000413605.2
|
CDK19
|
cyclin-dependent kinase 19 |
| chr12_-_53343560 | 0.02 |
ENST00000548998.1
|
KRT8
|
keratin 8 |
| chr2_-_220264703 | 0.02 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr5_+_78532003 | 0.02 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr5_+_65222500 | 0.02 |
ENST00000511297.1
ENST00000506030.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr4_-_140223670 | 0.02 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr18_-_53068911 | 0.02 |
ENST00000537856.3
|
TCF4
|
transcription factor 4 |
| chr10_+_111985837 | 0.02 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr11_+_71791359 | 0.02 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr15_+_42697018 | 0.02 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr6_+_22221010 | 0.02 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr5_+_162932554 | 0.02 |
ENST00000321757.6
ENST00000421814.2 ENST00000518095.1 |
MAT2B
|
methionine adenosyltransferase II, beta |
| chr12_-_58240470 | 0.02 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr19_-_36523709 | 0.02 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr4_+_146403912 | 0.02 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr15_+_72978539 | 0.02 |
ENST00000539603.1
ENST00000569338.1 |
BBS4
|
Bardet-Biedl syndrome 4 |
| chr6_+_32121908 | 0.02 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr14_+_88851874 | 0.02 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chrX_-_47863348 | 0.02 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr20_+_9049682 | 0.02 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr10_-_4285923 | 0.02 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr6_-_112575687 | 0.02 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr14_+_105267250 | 0.02 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr22_-_30198075 | 0.02 |
ENST00000411532.1
|
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr12_-_53343633 | 0.02 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr6_-_112575912 | 0.02 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr3_+_136676851 | 0.02 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr4_+_79567057 | 0.02 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr6_+_43027595 | 0.02 |
ENST00000259708.3
ENST00000472792.1 ENST00000479388.1 ENST00000460283.1 ENST00000394056.2 |
KLC4
|
kinesin light chain 4 |
| chr21_+_17443434 | 0.02 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr18_-_74844727 | 0.02 |
ENST00000355994.2
ENST00000579129.1 |
MBP
|
myelin basic protein |
| chr22_-_39268308 | 0.02 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr1_-_1356628 | 0.02 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr2_-_37193606 | 0.02 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr3_-_185826718 | 0.02 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr14_+_50234827 | 0.02 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr20_+_45338126 | 0.02 |
ENST00000359271.2
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr15_+_57540230 | 0.02 |
ENST00000559703.1
|
TCF12
|
transcription factor 12 |
| chr17_+_58755184 | 0.02 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr17_-_40134339 | 0.02 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr15_+_72978521 | 0.02 |
ENST00000542334.1
ENST00000268057.4 |
BBS4
|
Bardet-Biedl syndrome 4 |
| chr5_+_140734570 | 0.02 |
ENST00000571252.1
|
PCDHGA4
|
protocadherin gamma subfamily A, 4 |
| chr9_-_28026318 | 0.02 |
ENST00000308675.3
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr4_+_172734548 | 0.02 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr16_+_28763108 | 0.02 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr2_+_61404545 | 0.01 |
ENST00000357022.2
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
| chr8_-_70747205 | 0.01 |
ENST00000260126.4
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr4_+_166794383 | 0.01 |
ENST00000061240.2
ENST00000507499.1 |
TLL1
|
tolloid-like 1 |
| chr8_+_31497271 | 0.01 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr8_+_38243821 | 0.01 |
ENST00000519476.2
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr11_+_67374323 | 0.01 |
ENST00000322776.6
ENST00000532303.1 ENST00000532244.1 ENST00000528328.1 ENST00000529927.1 ENST00000532343.1 ENST00000415352.2 ENST00000533075.1 ENST00000529867.1 ENST00000530638.1 |
NDUFV1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa |
| chr10_-_103578162 | 0.01 |
ENST00000361464.3
ENST00000357797.5 ENST00000370094.3 |
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr5_-_59064458 | 0.01 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_41725140 | 0.01 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr4_-_82393052 | 0.01 |
ENST00000335927.7
ENST00000504863.1 ENST00000264400.2 |
RASGEF1B
|
RasGEF domain family, member 1B |
| chr20_-_62582475 | 0.01 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr10_+_52751010 | 0.01 |
ENST00000373985.1
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr19_-_49016418 | 0.01 |
ENST00000270238.3
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr12_-_71551652 | 0.01 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr10_-_103578182 | 0.01 |
ENST00000439817.1
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr2_-_172750733 | 0.01 |
ENST00000392592.4
ENST00000422440.2 |
SLC25A12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr12_-_21928515 | 0.01 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr19_+_50380682 | 0.01 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr21_+_17443521 | 0.01 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr14_+_38033252 | 0.01 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr17_+_58677539 | 0.01 |
ENST00000305921.3
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr7_-_144435985 | 0.01 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr5_-_19988288 | 0.01 |
ENST00000502796.1
ENST00000511273.1 |
CDH18
|
cadherin 18, type 2 |
| chr7_-_84121858 | 0.01 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr6_+_32121789 | 0.01 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr1_+_236305826 | 0.01 |
ENST00000366592.3
ENST00000366591.4 |
GPR137B
|
G protein-coupled receptor 137B |
| chr15_+_28624878 | 0.01 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr5_-_39424961 | 0.01 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr20_-_52645231 | 0.01 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr4_-_99851766 | 0.01 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr13_-_36920420 | 0.01 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr12_-_96390063 | 0.01 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr4_-_152149033 | 0.01 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXF2_FOXJ1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |