Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
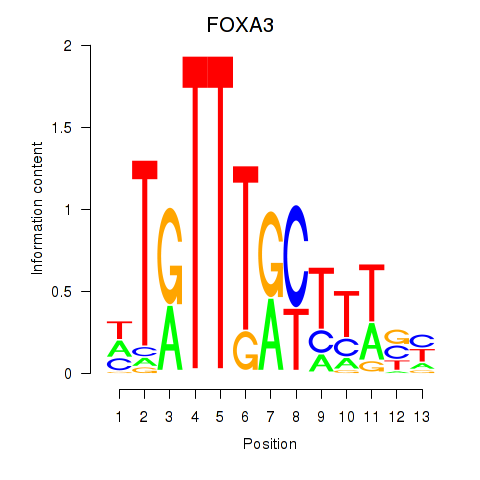
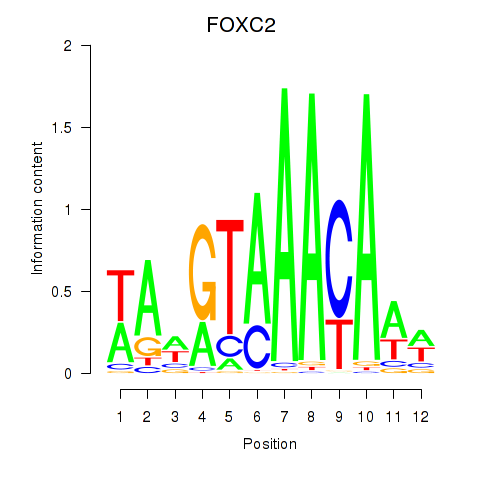
Results for FOXA3_FOXC2
Z-value: 0.62
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA3
|
ENSG00000170608.2 | forkhead box A3 |
|
FOXC2
|
ENSG00000176692.4 | forkhead box C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC2 | hg19_v2_chr16_+_86600857_86600921 | -0.09 | 8.7e-01 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_106415457 | 0.64 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr2_-_207078154 | 0.28 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr2_-_207078086 | 0.27 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr11_+_27076764 | 0.27 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr17_+_72426891 | 0.22 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr15_-_76352069 | 0.22 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr11_+_65657875 | 0.22 |
ENST00000312579.2
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr1_+_239882842 | 0.21 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr16_+_77233294 | 0.21 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr1_-_57431679 | 0.21 |
ENST00000371237.4
ENST00000535057.1 ENST00000543257.1 |
C8B
|
complement component 8, beta polypeptide |
| chr2_-_192711968 | 0.21 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr11_+_34999328 | 0.20 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr21_+_17791648 | 0.20 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_+_75174180 | 0.20 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chrX_-_15402498 | 0.20 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr3_-_193096600 | 0.19 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr10_+_124739964 | 0.19 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr13_-_86373536 | 0.19 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr2_+_171640291 | 0.19 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr7_+_134832808 | 0.19 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr19_+_42806250 | 0.19 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr14_-_31926623 | 0.18 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr2_-_175711133 | 0.17 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr12_-_78934441 | 0.17 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chr18_-_25616519 | 0.16 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr13_+_50589390 | 0.16 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr4_-_87028478 | 0.15 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_+_17997763 | 0.15 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr17_+_79953310 | 0.15 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr19_+_54368001 | 0.14 |
ENST00000422045.1
|
AC008440.10
|
AC008440.10 |
| chr17_+_72427477 | 0.14 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr7_-_117512264 | 0.14 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr7_+_142829162 | 0.13 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr11_+_65851443 | 0.13 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr8_+_97597148 | 0.13 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr4_-_186696561 | 0.12 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_8924837 | 0.12 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr8_+_67579807 | 0.12 |
ENST00000519289.1
ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3
C8orf44
|
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr3_-_139195350 | 0.12 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr3_-_107941209 | 0.12 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr14_-_36988882 | 0.12 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr14_-_31926701 | 0.11 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr5_+_169011033 | 0.11 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr17_-_65235916 | 0.11 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr12_-_92536433 | 0.11 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr2_-_188312971 | 0.11 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr16_-_4665023 | 0.11 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr12_-_62585203 | 0.11 |
ENST00000551449.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr15_-_59041954 | 0.11 |
ENST00000439637.1
ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr11_-_16419067 | 0.10 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr13_-_40924439 | 0.10 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr8_-_95449155 | 0.10 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr21_-_31588338 | 0.10 |
ENST00000286809.1
|
CLDN8
|
claudin 8 |
| chr7_+_129906660 | 0.10 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr16_-_4664860 | 0.10 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr19_+_782755 | 0.10 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr1_-_179851611 | 0.10 |
ENST00000610272.1
|
RP11-533E19.7
|
RP11-533E19.7 |
| chr17_+_61151306 | 0.10 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr5_-_24645078 | 0.10 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr12_+_28605426 | 0.10 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_172745006 | 0.09 |
ENST00000432694.2
|
RP1-15D23.2
|
RP1-15D23.2 |
| chr4_+_86396265 | 0.09 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr14_-_25479811 | 0.09 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr5_+_81601166 | 0.09 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr17_-_29641084 | 0.09 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr17_-_29641104 | 0.09 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr1_+_174933899 | 0.09 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_-_94789663 | 0.09 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr4_+_57138437 | 0.09 |
ENST00000504228.1
ENST00000541073.1 |
KIAA1211
|
KIAA1211 |
| chr10_-_21186144 | 0.09 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr14_+_74551650 | 0.09 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr9_+_67977438 | 0.09 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr17_-_39684550 | 0.09 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr17_-_36358166 | 0.09 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr20_+_9146969 | 0.09 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr6_+_101846664 | 0.09 |
ENST00000421544.1
ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr10_+_71561630 | 0.08 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr14_-_23526739 | 0.08 |
ENST00000397359.3
ENST00000487137.2 |
CDH24
|
cadherin 24, type 2 |
| chr4_+_110769258 | 0.08 |
ENST00000594814.1
|
LRIT3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chrX_-_80457385 | 0.08 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr3_+_177159695 | 0.08 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr4_-_140223614 | 0.08 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr22_-_39268308 | 0.08 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr1_-_54405773 | 0.08 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr10_-_4285835 | 0.07 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr20_+_9494987 | 0.07 |
ENST00000427562.2
ENST00000246070.2 |
LAMP5
|
lysosomal-associated membrane protein family, member 5 |
| chr10_+_71562180 | 0.07 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_+_68592305 | 0.07 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr10_-_46030841 | 0.07 |
ENST00000453424.2
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr12_-_7596735 | 0.07 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr21_-_31588365 | 0.07 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr17_+_4613918 | 0.07 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr3_+_177159744 | 0.07 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr6_+_142623758 | 0.07 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr17_-_202579 | 0.07 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr10_+_70847852 | 0.07 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr7_+_77469439 | 0.07 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr12_+_72080253 | 0.07 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr1_+_87797351 | 0.07 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr11_-_82708435 | 0.07 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr1_+_84609944 | 0.07 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_-_1356719 | 0.06 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr9_-_28670283 | 0.06 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr7_+_151791095 | 0.06 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr15_+_71185148 | 0.06 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr9_+_132044730 | 0.06 |
ENST00000455981.1
|
RP11-344B5.2
|
RP11-344B5.2 |
| chr9_-_75567962 | 0.06 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr15_+_96869165 | 0.06 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr10_+_71561649 | 0.06 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr10_-_4285923 | 0.06 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr17_-_26733604 | 0.06 |
ENST00000584426.1
ENST00000584995.1 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr1_-_1356628 | 0.06 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr12_+_48099858 | 0.06 |
ENST00000547799.1
|
RP1-197B17.3
|
RP1-197B17.3 |
| chr10_-_31288398 | 0.06 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr14_+_56585048 | 0.06 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr14_+_37126765 | 0.06 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chrX_+_107288280 | 0.05 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr9_+_71986182 | 0.05 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr9_-_4299874 | 0.05 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr14_-_36990354 | 0.05 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr13_+_95364963 | 0.05 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr16_-_48281305 | 0.05 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr17_+_4613776 | 0.05 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr12_+_40787194 | 0.05 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr1_-_207095324 | 0.05 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr11_-_116708302 | 0.05 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr22_+_46449674 | 0.05 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr9_+_131062367 | 0.05 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr17_-_6947225 | 0.05 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr8_+_52730143 | 0.05 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr2_-_21266816 | 0.05 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr6_-_26056695 | 0.05 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr4_-_140223670 | 0.05 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr21_-_36259445 | 0.05 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr4_-_153303658 | 0.05 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr10_+_95848824 | 0.05 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr17_+_74372662 | 0.05 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr2_+_175260514 | 0.05 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr12_+_59989791 | 0.05 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr16_+_20775358 | 0.05 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr1_+_101361782 | 0.05 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr5_+_137673200 | 0.05 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr5_+_95998673 | 0.05 |
ENST00000514845.1
|
CAST
|
calpastatin |
| chr10_-_46030787 | 0.05 |
ENST00000395769.2
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr21_+_17791838 | 0.05 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_-_108464321 | 0.05 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr2_-_220042825 | 0.04 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr4_+_147096837 | 0.04 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr4_+_184826418 | 0.04 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr17_-_41322332 | 0.04 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr2_-_106054952 | 0.04 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr11_-_83984231 | 0.04 |
ENST00000330014.6
ENST00000537455.1 ENST00000376106.3 ENST00000418306.2 ENST00000531015.1 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr8_-_71157595 | 0.04 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr6_-_122792919 | 0.04 |
ENST00000339697.4
|
SERINC1
|
serine incorporator 1 |
| chr1_-_177939348 | 0.04 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr4_+_166794383 | 0.04 |
ENST00000061240.2
ENST00000507499.1 |
TLL1
|
tolloid-like 1 |
| chr1_-_167522982 | 0.04 |
ENST00000370509.4
|
CREG1
|
cellular repressor of E1A-stimulated genes 1 |
| chr15_+_76016293 | 0.04 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr4_+_172734548 | 0.04 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr1_+_29563011 | 0.04 |
ENST00000345512.3
ENST00000373779.3 ENST00000356870.3 ENST00000323874.8 ENST00000428026.2 ENST00000460170.2 |
PTPRU
|
protein tyrosine phosphatase, receptor type, U |
| chr11_-_66112555 | 0.04 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr8_-_28243934 | 0.04 |
ENST00000521185.1
ENST00000520290.1 ENST00000344423.5 |
ZNF395
|
zinc finger protein 395 |
| chr4_+_89206076 | 0.04 |
ENST00000500009.2
|
RP11-10L7.1
|
RP11-10L7.1 |
| chr12_-_120765565 | 0.04 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr2_-_32490859 | 0.04 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr17_-_39140549 | 0.04 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr3_-_107941230 | 0.04 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr4_-_186570679 | 0.04 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_65222500 | 0.04 |
ENST00000511297.1
ENST00000506030.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr1_+_179851893 | 0.04 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr14_-_74551096 | 0.04 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr1_+_40974431 | 0.04 |
ENST00000296380.4
ENST00000432259.1 ENST00000418186.1 |
EXO5
|
exonuclease 5 |
| chr16_+_19619083 | 0.04 |
ENST00000538552.1
|
C16orf62
|
chromosome 16 open reading frame 62 |
| chr19_+_47421933 | 0.03 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr9_+_4839762 | 0.03 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr20_+_9049742 | 0.03 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr1_-_146696901 | 0.03 |
ENST00000369272.3
ENST00000441068.2 |
FMO5
|
flavin containing monooxygenase 5 |
| chr2_-_172290482 | 0.03 |
ENST00000442541.1
ENST00000392599.2 ENST00000375258.4 |
METTL8
|
methyltransferase like 8 |
| chr2_-_65593784 | 0.03 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr18_-_53070913 | 0.03 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr20_+_44519948 | 0.03 |
ENST00000354880.5
ENST00000191018.5 |
CTSA
|
cathepsin A |
| chr20_-_43150601 | 0.03 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr19_+_50380682 | 0.03 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr12_+_21525818 | 0.03 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr20_+_31870927 | 0.03 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr14_-_74551172 | 0.03 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr17_-_26697304 | 0.03 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr1_+_73771844 | 0.03 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr12_-_21928515 | 0.03 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr20_+_44520009 | 0.03 |
ENST00000607482.1
ENST00000372459.2 |
CTSA
|
cathepsin A |
| chrX_-_47863348 | 0.03 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr2_+_220042933 | 0.03 |
ENST00000430297.2
|
FAM134A
|
family with sequence similarity 134, member A |
| chr11_-_108464465 | 0.03 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr1_-_43855444 | 0.03 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr14_+_105267250 | 0.03 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr15_+_38964048 | 0.03 |
ENST00000560203.1
ENST00000557946.1 |
RP11-275I4.2
|
RP11-275I4.2 |
| chr1_+_172389821 | 0.03 |
ENST00000367727.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr6_-_159466042 | 0.03 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr7_+_116654958 | 0.03 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr6_-_112575912 | 0.03 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.0 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.0 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |