Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
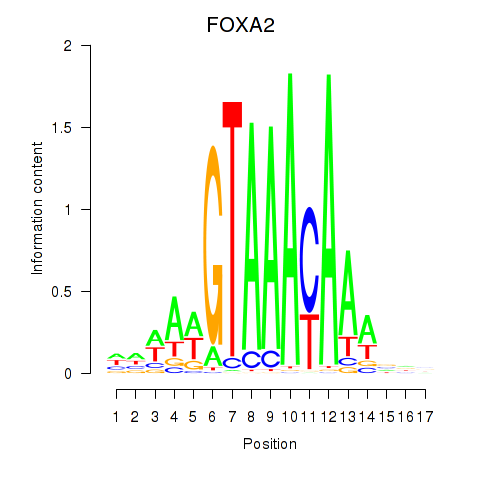
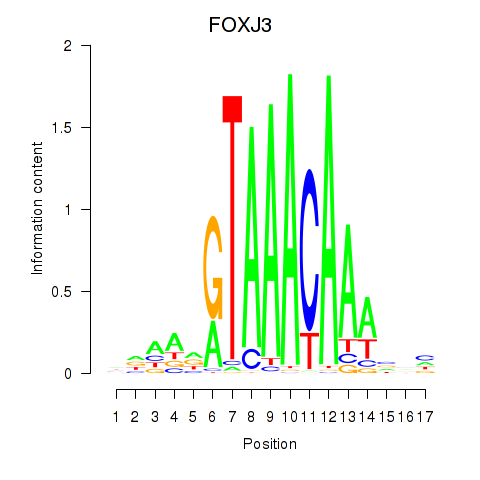
Results for FOXA2_FOXJ3
Z-value: 0.71
Transcription factors associated with FOXA2_FOXJ3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA2
|
ENSG00000125798.10 | forkhead box A2 |
|
FOXJ3
|
ENSG00000198815.4 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA2 | hg19_v2_chr20_-_22565101_22565223 | -0.34 | 5.1e-01 | Click! |
| FOXJ3 | hg19_v2_chr1_-_42800860_42800912 | 0.05 | 9.3e-01 | Click! |
Activity profile of FOXA2_FOXJ3 motif
Sorted Z-values of FOXA2_FOXJ3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_72129806 | 0.73 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr2_+_58655461 | 0.57 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr10_-_4285835 | 0.54 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr6_-_152623231 | 0.52 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_246729544 | 0.34 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr12_-_12714006 | 0.33 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr16_+_28857916 | 0.32 |
ENST00000563591.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr5_+_61708488 | 0.31 |
ENST00000505902.1
|
IPO11
|
importin 11 |
| chr12_-_15038779 | 0.29 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr6_+_122793058 | 0.27 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr10_-_4285923 | 0.27 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr12_-_106480587 | 0.26 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr6_+_161123270 | 0.25 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr12_-_12714025 | 0.24 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr12_-_92539614 | 0.24 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr1_-_161193349 | 0.24 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr17_-_57229155 | 0.24 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr6_+_26217159 | 0.24 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr19_+_782755 | 0.24 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr2_+_88047606 | 0.23 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr3_-_48956818 | 0.23 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr19_+_16607122 | 0.23 |
ENST00000221671.3
ENST00000594035.1 ENST00000599550.1 ENST00000594813.1 |
C19orf44
|
chromosome 19 open reading frame 44 |
| chr17_+_67590125 | 0.23 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr12_+_57998400 | 0.23 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr2_+_162165038 | 0.20 |
ENST00000437630.1
|
PSMD14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr18_+_68002675 | 0.20 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chrX_+_9431324 | 0.20 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr4_+_104346194 | 0.20 |
ENST00000510200.1
|
RP11-328K4.1
|
RP11-328K4.1 |
| chr16_-_21442874 | 0.19 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr1_+_92545862 | 0.19 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr19_-_8373173 | 0.19 |
ENST00000537716.2
ENST00000301458.5 |
CD320
|
CD320 molecule |
| chr12_+_59989918 | 0.19 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_-_135916073 | 0.19 |
ENST00000481989.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr10_+_33271469 | 0.19 |
ENST00000414157.1
|
RP11-462L8.1
|
RP11-462L8.1 |
| chr2_+_66918558 | 0.18 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr12_+_28605426 | 0.18 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr2_+_143635222 | 0.17 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr17_+_41561317 | 0.16 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr17_-_6915616 | 0.16 |
ENST00000575889.1
|
AC027763.2
|
Uncharacterized protein |
| chr5_-_150460539 | 0.16 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_-_114430169 | 0.15 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr2_+_143635067 | 0.15 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr1_-_207095324 | 0.15 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr1_+_66796401 | 0.15 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr1_+_150122034 | 0.15 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr8_+_103540983 | 0.15 |
ENST00000523572.1
|
KB-1980E6.3
|
Uncharacterized protein |
| chr16_-_88772670 | 0.14 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr1_-_203273676 | 0.13 |
ENST00000425698.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr17_-_26662440 | 0.13 |
ENST00000578122.1
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr19_-_52307357 | 0.13 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr5_+_136070614 | 0.13 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr12_+_49297899 | 0.13 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr4_+_154622652 | 0.13 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr17_+_26662679 | 0.13 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr5_-_115872124 | 0.13 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr12_-_7596735 | 0.12 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr15_+_90735145 | 0.12 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr15_+_64680003 | 0.12 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr3_+_171561127 | 0.12 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr16_+_22517166 | 0.12 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr2_+_162272605 | 0.12 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr13_-_99910673 | 0.12 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr2_+_12857043 | 0.11 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr1_-_43855444 | 0.11 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr6_-_26216872 | 0.11 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr8_+_67104323 | 0.11 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr7_+_106809406 | 0.11 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr4_+_117220016 | 0.11 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr19_+_36132631 | 0.11 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr7_+_76107444 | 0.11 |
ENST00000435861.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr15_-_40398812 | 0.10 |
ENST00000561360.1
|
BMF
|
Bcl2 modifying factor |
| chr1_-_89736434 | 0.10 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr11_+_65851443 | 0.10 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr7_+_76090993 | 0.10 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr15_+_90792760 | 0.10 |
ENST00000339615.5
ENST00000438251.1 |
TTLL13
|
tubulin tyrosine ligase-like family, member 13 |
| chr20_+_44035847 | 0.10 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr12_-_49582978 | 0.10 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr10_+_78078088 | 0.10 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr14_+_58894404 | 0.10 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr6_-_2751146 | 0.10 |
ENST00000268446.5
ENST00000274643.7 |
MYLK4
|
myosin light chain kinase family, member 4 |
| chr7_-_139876734 | 0.09 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr1_+_203765437 | 0.09 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr1_-_3566627 | 0.09 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr11_-_92930556 | 0.09 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr16_+_22518495 | 0.09 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr6_-_117747015 | 0.09 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr4_-_80994619 | 0.09 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr3_-_169487617 | 0.09 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr17_+_26662730 | 0.09 |
ENST00000226225.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr9_-_20382446 | 0.09 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr14_+_102276192 | 0.09 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr14_+_100070869 | 0.09 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr17_-_65235916 | 0.09 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr11_-_7904464 | 0.09 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr7_+_6617039 | 0.09 |
ENST00000405731.3
ENST00000396713.2 ENST00000396707.2 ENST00000335965.6 ENST00000396709.1 ENST00000483589.1 ENST00000396706.2 |
ZDHHC4
|
zinc finger, DHHC-type containing 4 |
| chr16_-_28857677 | 0.09 |
ENST00000313511.3
|
TUFM
|
Tu translation elongation factor, mitochondrial |
| chr1_-_114429997 | 0.08 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr8_+_21777159 | 0.08 |
ENST00000434536.1
ENST00000252512.9 |
XPO7
|
exportin 7 |
| chr19_+_3880581 | 0.08 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr9_-_86432547 | 0.08 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr9_+_112852477 | 0.08 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr8_+_75512010 | 0.08 |
ENST00000518190.1
ENST00000523118.1 |
RP11-758M4.1
|
Uncharacterized protein |
| chr19_+_12721760 | 0.08 |
ENST00000600752.1
ENST00000540038.1 |
ZNF791
|
zinc finger protein 791 |
| chr17_+_41363854 | 0.08 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr21_-_16374688 | 0.08 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr5_-_13944652 | 0.08 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr11_-_102651343 | 0.08 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr1_-_207095212 | 0.08 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr6_+_12958137 | 0.08 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr8_-_101321584 | 0.08 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr17_-_30228678 | 0.08 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr10_-_69597828 | 0.08 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr17_+_7942424 | 0.08 |
ENST00000573359.1
|
ALOX15B
|
arachidonate 15-lipoxygenase, type B |
| chr19_+_35899569 | 0.08 |
ENST00000600405.1
|
AC002511.1
|
AC002511.1 |
| chr11_+_34643600 | 0.08 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr17_-_71223839 | 0.07 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr7_-_16840820 | 0.07 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr9_-_136933615 | 0.07 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr12_+_120740119 | 0.07 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr16_+_22516172 | 0.07 |
ENST00000543407.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr17_+_685513 | 0.07 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
| chr10_+_124739911 | 0.07 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr5_-_96209315 | 0.07 |
ENST00000504056.1
|
CTD-2260A17.2
|
Uncharacterized protein |
| chr22_-_22295029 | 0.07 |
ENST00000445205.1
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr4_+_28437071 | 0.07 |
ENST00000509416.1
|
RP11-123O22.1
|
RP11-123O22.1 |
| chr21_+_34602377 | 0.07 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr12_-_76879852 | 0.07 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr3_+_142315294 | 0.07 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr2_+_171640291 | 0.07 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr17_-_72772425 | 0.07 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr21_-_46293644 | 0.07 |
ENST00000330938.3
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein |
| chr5_+_147774275 | 0.07 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr5_+_137673945 | 0.07 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr16_-_279405 | 0.07 |
ENST00000430864.1
ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L
|
LUC7-like (S. cerevisiae) |
| chr2_+_149402989 | 0.07 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr6_-_88032795 | 0.07 |
ENST00000296882.3
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr17_-_6915646 | 0.07 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chrX_-_65259914 | 0.07 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr3_+_172468749 | 0.07 |
ENST00000366254.2
ENST00000415665.1 ENST00000438041.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr14_+_102606181 | 0.07 |
ENST00000335263.5
ENST00000322340.5 ENST00000424963.2 ENST00000342702.3 ENST00000556807.1 ENST00000499851.2 ENST00000558567.1 ENST00000299135.6 ENST00000454394.2 ENST00000556511.2 |
WDR20
|
WD repeat domain 20 |
| chr17_-_72772462 | 0.07 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr3_+_109128961 | 0.07 |
ENST00000489670.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr12_+_2986359 | 0.07 |
ENST00000538636.1
ENST00000461997.2 ENST00000489288.2 ENST00000366285.2 ENST00000538700.1 |
RHNO1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr8_+_86999516 | 0.06 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr1_+_45140360 | 0.06 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr14_+_97263641 | 0.06 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr8_+_103991013 | 0.06 |
ENST00000517983.1
|
KB-1507C5.4
|
KB-1507C5.4 |
| chr2_+_191513959 | 0.06 |
ENST00000337386.5
ENST00000357215.5 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr12_+_97306295 | 0.06 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr9_-_35812140 | 0.06 |
ENST00000497810.1
ENST00000396638.2 ENST00000484764.1 |
SPAG8
|
sperm associated antigen 8 |
| chr17_+_77030267 | 0.06 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_-_42931567 | 0.06 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr17_+_26662597 | 0.06 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr4_-_21950356 | 0.06 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr19_+_6464243 | 0.06 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr10_+_97759848 | 0.06 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr19_-_4831701 | 0.06 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr5_+_176853669 | 0.06 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr8_+_11666649 | 0.06 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr8_-_33424636 | 0.06 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr19_+_36486078 | 0.06 |
ENST00000378887.2
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1 |
| chr14_-_74551172 | 0.06 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr5_-_98262240 | 0.06 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr1_+_41448820 | 0.06 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr1_+_246729815 | 0.06 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr16_+_30064411 | 0.06 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr8_+_101349823 | 0.06 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr15_+_35270552 | 0.06 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr1_+_163039143 | 0.06 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr16_+_30064444 | 0.06 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_-_156272872 | 0.06 |
ENST00000476217.1
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr4_+_86748898 | 0.06 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr16_+_84801852 | 0.06 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr8_+_67344710 | 0.06 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr7_+_90338547 | 0.06 |
ENST00000446790.1
|
CDK14
|
cyclin-dependent kinase 14 |
| chr6_+_26251835 | 0.06 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr6_-_131211534 | 0.06 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_-_60632162 | 0.06 |
ENST00000557185.1
|
DHRS7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr19_+_18111927 | 0.06 |
ENST00000379656.3
|
ARRDC2
|
arrestin domain containing 2 |
| chr4_-_80994471 | 0.06 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr19_+_15160130 | 0.06 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr3_-_187455680 | 0.06 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr11_-_61647935 | 0.06 |
ENST00000531956.1
|
FADS3
|
fatty acid desaturase 3 |
| chr21_+_34602200 | 0.06 |
ENST00000382264.3
ENST00000382241.3 ENST00000404220.3 ENST00000342136.4 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr5_+_179135246 | 0.06 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr10_+_90521163 | 0.05 |
ENST00000404459.1
|
LIPN
|
lipase, family member N |
| chr1_+_241695670 | 0.05 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr7_+_74191613 | 0.05 |
ENST00000442021.2
|
NCF1
|
neutrophil cytosolic factor 1 |
| chr19_+_43325937 | 0.05 |
ENST00000425668.1
|
AC004603.4
|
AC004603.4 |
| chr5_-_88119580 | 0.05 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr7_+_26332645 | 0.05 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr8_-_93978309 | 0.05 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr15_-_43559055 | 0.05 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr10_-_101825151 | 0.05 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr2_-_240230890 | 0.05 |
ENST00000446876.1
|
HDAC4
|
histone deacetylase 4 |
| chr14_-_58893832 | 0.05 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_+_84906338 | 0.05 |
ENST00000512109.1
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr12_-_75905374 | 0.05 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr9_-_35812236 | 0.05 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr22_-_22337154 | 0.05 |
ENST00000413067.2
ENST00000437929.1 ENST00000456075.1 ENST00000434517.1 ENST00000424393.1 ENST00000449704.1 ENST00000437103.1 |
TOP3B
|
topoisomerase (DNA) III beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA2_FOXJ3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.1 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.5 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:1903217 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0039008 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.0 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.0 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 0.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |