Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
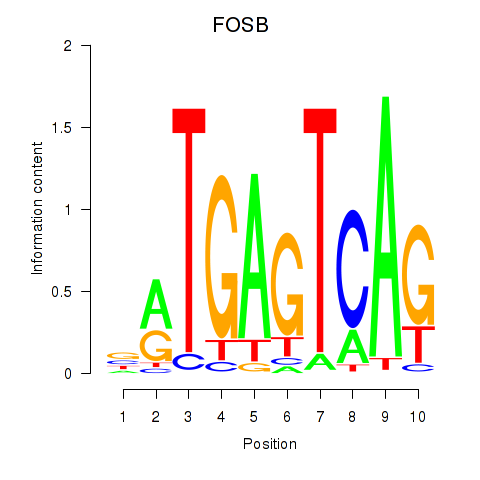
Results for FOSB
Z-value: 0.48
Transcription factors associated with FOSB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSB
|
ENSG00000125740.9 | FosB proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSB | hg19_v2_chr19_+_45973120_45973171 | 0.72 | 1.1e-01 | Click! |
Activity profile of FOSB motif
Sorted Z-values of FOSB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_51863853 | 0.71 |
ENST00000556762.1
|
RP11-255G12.3
|
RP11-255G12.3 |
| chr5_+_147582348 | 0.50 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr10_+_88780049 | 0.46 |
ENST00000343959.4
|
FAM25A
|
family with sequence similarity 25, member A |
| chr4_+_74606223 | 0.38 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr1_-_153029980 | 0.33 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr8_+_125283924 | 0.30 |
ENST00000517482.1
|
RP11-383J24.1
|
RP11-383J24.1 |
| chr19_-_46627914 | 0.29 |
ENST00000341415.2
|
IGFL3
|
IGF-like family member 3 |
| chr10_+_1102303 | 0.20 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr5_+_147582387 | 0.20 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr17_+_4853442 | 0.20 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr16_+_89988259 | 0.19 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr17_+_65375082 | 0.19 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr20_+_44637526 | 0.18 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr16_-_57935277 | 0.18 |
ENST00000565942.1
|
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr4_-_89079817 | 0.17 |
ENST00000505480.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr1_+_152881014 | 0.17 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr19_-_44174330 | 0.16 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr11_-_102714534 | 0.16 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr8_+_99076509 | 0.16 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr11_-_65667884 | 0.16 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr4_-_69083720 | 0.16 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr5_+_53751445 | 0.15 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr9_+_44867571 | 0.15 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr17_+_46970134 | 0.15 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr2_+_152214098 | 0.15 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr2_+_87754989 | 0.14 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr19_-_44174305 | 0.14 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr10_-_47181681 | 0.14 |
ENST00000452267.1
|
FAM25B
|
family with sequence similarity 25, member B |
| chr11_-_102668879 | 0.14 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr10_+_48247669 | 0.13 |
ENST00000457620.1
|
FAM25G
|
family with sequence similarity 25, member G |
| chr9_-_34662651 | 0.13 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr12_+_7023735 | 0.13 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr2_+_135596180 | 0.13 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_+_124997766 | 0.13 |
ENST00000543970.1
|
RP11-83B20.1
|
RP11-83B20.1 |
| chr17_+_46970127 | 0.13 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr22_+_37959647 | 0.13 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr2_+_208104497 | 0.12 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr11_-_65667997 | 0.12 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr17_-_18908040 | 0.12 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr10_+_75668916 | 0.12 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr2_-_219858123 | 0.12 |
ENST00000453769.1
ENST00000295728.2 ENST00000392096.2 |
CRYBA2
|
crystallin, beta A2 |
| chr2_-_218766698 | 0.11 |
ENST00000439083.1
|
TNS1
|
tensin 1 |
| chr2_+_102618428 | 0.11 |
ENST00000457817.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chr19_-_6057282 | 0.11 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr3_+_14474178 | 0.11 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr7_+_28448995 | 0.11 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr16_+_56782118 | 0.11 |
ENST00000566678.1
|
NUP93
|
nucleoporin 93kDa |
| chr21_-_35340759 | 0.11 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr1_+_165796753 | 0.11 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr19_-_18995029 | 0.11 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr14_-_75083313 | 0.11 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr8_-_37411648 | 0.11 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr8_-_38386175 | 0.11 |
ENST00000437935.2
ENST00000358138.1 |
C8orf86
|
chromosome 8 open reading frame 86 |
| chr5_-_43412418 | 0.10 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr1_-_95007193 | 0.10 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr11_-_2924970 | 0.10 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr12_+_7023491 | 0.10 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr14_+_73525144 | 0.10 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr12_-_15114658 | 0.10 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_-_45272951 | 0.10 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr3_-_131756559 | 0.10 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr8_+_87111059 | 0.10 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr1_-_235116495 | 0.09 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr15_-_60690163 | 0.09 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr11_-_64527425 | 0.09 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr6_-_30685214 | 0.09 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr14_-_75536182 | 0.09 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr3_-_141747950 | 0.09 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_149293990 | 0.09 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr18_+_21452804 | 0.09 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr17_+_35851570 | 0.09 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr12_-_95510743 | 0.09 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr17_+_9745786 | 0.09 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr1_-_115880852 | 0.09 |
ENST00000369512.2
|
NGF
|
nerve growth factor (beta polypeptide) |
| chr1_+_44584522 | 0.09 |
ENST00000372299.3
|
KLF17
|
Kruppel-like factor 17 |
| chr22_+_39916558 | 0.09 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr17_+_30771279 | 0.09 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr12_-_54813229 | 0.09 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr3_-_141747439 | 0.09 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr15_+_45938079 | 0.09 |
ENST00000561493.1
|
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr18_+_21452964 | 0.09 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr14_+_73525229 | 0.09 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr11_+_10477733 | 0.09 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr2_+_208104351 | 0.09 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr13_-_41768654 | 0.09 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr2_-_11606275 | 0.08 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr6_+_33043703 | 0.08 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr20_+_21686290 | 0.08 |
ENST00000398485.2
|
PAX1
|
paired box 1 |
| chr11_-_10920838 | 0.08 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr17_+_46970178 | 0.08 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr10_-_111713633 | 0.08 |
ENST00000538668.1
ENST00000369657.1 ENST00000369655.1 |
RP11-451M19.3
|
Uncharacterized protein |
| chr4_+_22999152 | 0.08 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr2_-_11605966 | 0.08 |
ENST00000307236.4
ENST00000542100.1 ENST00000546212.1 |
E2F6
|
E2F transcription factor 6 |
| chr4_+_76481258 | 0.08 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr16_-_30125177 | 0.08 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr4_+_71588372 | 0.08 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr19_-_35981358 | 0.08 |
ENST00000484218.2
ENST00000338897.3 |
KRTDAP
|
keratinocyte differentiation-associated protein |
| chr3_-_52869205 | 0.07 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr6_+_292051 | 0.07 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr11_-_102826434 | 0.07 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr8_-_133772870 | 0.07 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr2_+_135596106 | 0.07 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr17_+_28256874 | 0.07 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr12_-_15114603 | 0.07 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr11_+_59522837 | 0.07 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr6_-_35888905 | 0.07 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr20_+_48429356 | 0.07 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr1_+_110026544 | 0.07 |
ENST00000369870.3
|
ATXN7L2
|
ataxin 7-like 2 |
| chr5_+_145316120 | 0.07 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr15_+_41062159 | 0.07 |
ENST00000344320.6
|
C15orf62
|
chromosome 15 open reading frame 62 |
| chr17_-_3595042 | 0.07 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr19_+_47104553 | 0.07 |
ENST00000598871.1
ENST00000594523.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr14_-_22005197 | 0.07 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_+_155538093 | 0.07 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr1_+_26606608 | 0.07 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr14_+_73525265 | 0.07 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr9_-_35112376 | 0.07 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr15_+_67418047 | 0.07 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr6_-_31704282 | 0.06 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr18_+_32290218 | 0.06 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr2_-_136633940 | 0.06 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr8_-_110988070 | 0.06 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr6_-_24877490 | 0.06 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr17_-_8021710 | 0.06 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr9_+_34992846 | 0.06 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr4_+_84457250 | 0.06 |
ENST00000395226.2
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr1_+_27719148 | 0.06 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr16_-_1429627 | 0.06 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr15_+_77287426 | 0.06 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr18_+_9885760 | 0.06 |
ENST00000536353.2
ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr19_+_38880695 | 0.06 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr12_-_15114191 | 0.06 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr4_+_84457529 | 0.06 |
ENST00000264409.4
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr20_-_634000 | 0.06 |
ENST00000381962.3
|
SRXN1
|
sulfiredoxin 1 |
| chr13_+_76334498 | 0.06 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr7_+_116312411 | 0.05 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chrX_-_48901012 | 0.05 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr5_-_66942617 | 0.05 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr3_+_113465866 | 0.05 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr17_-_39769005 | 0.05 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr5_+_145317356 | 0.05 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr4_-_122085469 | 0.05 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr9_-_77502636 | 0.05 |
ENST00000449912.2
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr12_+_64846129 | 0.05 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr12_+_64845864 | 0.05 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr12_+_58176525 | 0.05 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr17_-_16918271 | 0.05 |
ENST00000562897.1
|
RP11-416I2.1
|
RP11-416I2.1 |
| chr12_-_15114492 | 0.05 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr4_+_71587669 | 0.05 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr16_+_31366455 | 0.05 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr19_+_38880252 | 0.05 |
ENST00000586301.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr9_+_140135665 | 0.05 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr7_+_30960915 | 0.05 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr15_+_22892663 | 0.05 |
ENST00000313077.7
ENST00000561274.1 ENST00000560848.1 |
CYFIP1
|
cytoplasmic FMR1 interacting protein 1 |
| chr6_-_31745037 | 0.05 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr5_+_66124590 | 0.05 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr20_+_48429233 | 0.05 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_+_7482785 | 0.05 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr22_-_32058166 | 0.04 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr17_+_37030127 | 0.04 |
ENST00000419929.1
|
LASP1
|
LIM and SH3 protein 1 |
| chr12_+_120972606 | 0.04 |
ENST00000413266.2
|
RNF10
|
ring finger protein 10 |
| chr12_-_49351228 | 0.04 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr5_-_141061777 | 0.04 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr13_-_67802549 | 0.04 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr11_-_64013288 | 0.04 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_-_65150103 | 0.04 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr20_-_48782639 | 0.04 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr1_-_21948906 | 0.04 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr8_+_70379072 | 0.04 |
ENST00000529134.1
|
SULF1
|
sulfatase 1 |
| chr2_-_238499131 | 0.04 |
ENST00000538644.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr11_-_8739383 | 0.04 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr11_-_68780824 | 0.04 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr2_+_16080659 | 0.04 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr19_+_6740888 | 0.04 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr1_-_84464780 | 0.04 |
ENST00000260505.8
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7 |
| chr2_+_87754887 | 0.04 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr22_-_32058416 | 0.04 |
ENST00000439502.2
|
PISD
|
phosphatidylserine decarboxylase |
| chr1_+_198189921 | 0.04 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr5_-_41794313 | 0.04 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr6_-_35888858 | 0.04 |
ENST00000507909.1
|
SRPK1
|
SRSF protein kinase 1 |
| chr14_+_31494672 | 0.04 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr5_+_38148582 | 0.04 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr17_+_7123125 | 0.04 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr1_-_9953295 | 0.04 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr19_+_47104493 | 0.04 |
ENST00000291295.9
ENST00000597743.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr5_-_141061759 | 0.04 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr5_+_140729649 | 0.03 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr9_+_131902346 | 0.03 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr3_-_98241713 | 0.03 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chr2_+_233562015 | 0.03 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr8_+_70476088 | 0.03 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chr7_-_130597935 | 0.03 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr17_-_18161870 | 0.03 |
ENST00000579294.1
ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII
|
flightless I homolog (Drosophila) |
| chr21_+_19617140 | 0.03 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr3_+_183903811 | 0.03 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr6_-_35888824 | 0.03 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr15_-_77363375 | 0.03 |
ENST00000559494.1
|
TSPAN3
|
tetraspanin 3 |
| chr12_-_51402984 | 0.03 |
ENST00000545993.2
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr15_-_77363513 | 0.03 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr11_-_71781096 | 0.03 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.5 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.2 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0044035 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.0 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.0 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.0 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.0 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |