Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
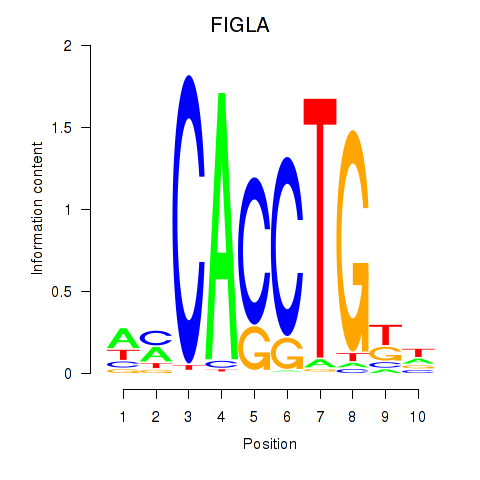
Results for FIGLA
Z-value: 0.47
Transcription factors associated with FIGLA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FIGLA
|
ENSG00000183733.6 | folliculogenesis specific bHLH transcription factor |
Activity profile of FIGLA motif
Sorted Z-values of FIGLA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_18391708 | 0.35 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr6_-_31697255 | 0.27 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr16_-_30006922 | 0.23 |
ENST00000564026.1
|
HIRIP3
|
HIRA interacting protein 3 |
| chr16_+_4845379 | 0.23 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr4_-_40516560 | 0.23 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr2_+_241375069 | 0.20 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr5_-_159739483 | 0.19 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr17_-_48133054 | 0.19 |
ENST00000499842.1
|
RP11-1094H24.4
|
RP11-1094H24.4 |
| chr6_+_31783291 | 0.18 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chrX_+_53078465 | 0.18 |
ENST00000375466.2
|
GPR173
|
G protein-coupled receptor 173 |
| chr6_+_31795506 | 0.18 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr16_+_30006997 | 0.17 |
ENST00000304516.7
|
INO80E
|
INO80 complex subunit E |
| chr1_-_219615984 | 0.17 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr6_-_31697563 | 0.16 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr19_+_55996565 | 0.16 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr16_+_30006615 | 0.16 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr7_-_120497178 | 0.16 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr1_+_10509971 | 0.15 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr3_+_32280159 | 0.15 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr3_+_49027771 | 0.15 |
ENST00000475629.1
ENST00000444213.1 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr15_+_89182156 | 0.15 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_+_81299370 | 0.14 |
ENST00000560091.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr17_+_74732889 | 0.14 |
ENST00000591864.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr4_+_6271558 | 0.14 |
ENST00000503569.1
ENST00000226760.1 |
WFS1
|
Wolfram syndrome 1 (wolframin) |
| chr17_+_27895609 | 0.14 |
ENST00000581411.2
ENST00000301057.7 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr12_-_53242770 | 0.14 |
ENST00000304620.4
ENST00000547110.1 |
KRT78
|
keratin 78 |
| chr17_+_73521763 | 0.13 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr17_+_39421591 | 0.13 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr11_+_22646739 | 0.13 |
ENST00000428556.2
|
AC103801.2
|
AC103801.2 |
| chr9_-_22009241 | 0.13 |
ENST00000380142.4
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr8_-_145018905 | 0.13 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr6_+_46761118 | 0.12 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr7_+_2687173 | 0.12 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr1_+_110754094 | 0.12 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr17_+_48133459 | 0.12 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr15_+_89182178 | 0.12 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr14_-_50999373 | 0.12 |
ENST00000554273.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr19_-_42916499 | 0.12 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr19_-_49118067 | 0.12 |
ENST00000593772.1
|
FAM83E
|
family with sequence similarity 83, member E |
| chr19_+_35739782 | 0.11 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739597 | 0.11 |
ENST00000361790.3
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_14017116 | 0.11 |
ENST00000589606.1
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr6_-_133055815 | 0.11 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr17_-_39538550 | 0.11 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr1_-_40367530 | 0.11 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr12_+_6419877 | 0.11 |
ENST00000536531.1
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr6_-_31938700 | 0.11 |
ENST00000495340.1
|
DXO
|
decapping exoribonuclease |
| chr20_-_22566089 | 0.11 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr6_-_36355513 | 0.11 |
ENST00000340181.4
ENST00000373737.4 |
ETV7
|
ets variant 7 |
| chr6_-_133055896 | 0.10 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr19_+_6464502 | 0.10 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr2_-_214013353 | 0.10 |
ENST00000451136.2
ENST00000421754.2 ENST00000374327.4 ENST00000413091.3 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr8_-_123706338 | 0.10 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr4_-_140223614 | 0.10 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr19_+_14017003 | 0.09 |
ENST00000318003.7
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr17_+_48133330 | 0.09 |
ENST00000544892.1
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr19_-_10230562 | 0.09 |
ENST00000587146.1
ENST00000588709.1 ENST00000253108.4 |
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr6_-_31550192 | 0.09 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr19_+_6464243 | 0.09 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr19_+_35739631 | 0.09 |
ENST00000602003.1
ENST00000360798.3 ENST00000354900.3 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr15_+_91411810 | 0.08 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_+_220436917 | 0.08 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr19_+_35739280 | 0.08 |
ENST00000602122.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_-_4902877 | 0.08 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr6_+_33172407 | 0.08 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr7_-_97881429 | 0.08 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr3_-_49851313 | 0.08 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr16_+_77225071 | 0.08 |
ENST00000439557.2
ENST00000545553.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr16_-_57513657 | 0.08 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr16_-_2260834 | 0.08 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chr16_-_103572 | 0.08 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr16_+_67233412 | 0.08 |
ENST00000477898.1
|
ELMO3
|
engulfment and cell motility 3 |
| chr14_-_37051798 | 0.08 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr3_-_50360192 | 0.08 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr6_+_7107830 | 0.08 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr3_-_178789220 | 0.08 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr6_-_33281979 | 0.08 |
ENST00000426633.2
ENST00000467025.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr2_+_101437487 | 0.08 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr15_-_45694380 | 0.08 |
ENST00000561148.1
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr4_-_108204904 | 0.08 |
ENST00000510463.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr14_-_107219365 | 0.07 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr7_+_140103842 | 0.07 |
ENST00000495590.1
ENST00000275874.5 ENST00000537763.1 |
RAB19
|
RAB19, member RAS oncogene family |
| chr3_-_50374869 | 0.07 |
ENST00000327761.3
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr2_-_27603582 | 0.07 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr7_-_8302298 | 0.07 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr8_+_145438870 | 0.07 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr19_-_46272106 | 0.07 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr15_-_45670717 | 0.07 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr17_-_46657473 | 0.07 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr16_+_88704978 | 0.07 |
ENST00000244241.4
|
IL17C
|
interleukin 17C |
| chr6_+_96463840 | 0.07 |
ENST00000302103.5
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase) |
| chr5_+_32712363 | 0.07 |
ENST00000507141.1
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr7_+_99699179 | 0.07 |
ENST00000438383.1
ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr7_-_11871815 | 0.07 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr7_-_27196267 | 0.07 |
ENST00000242159.3
|
HOXA7
|
homeobox A7 |
| chr12_-_80084333 | 0.07 |
ENST00000552637.1
|
PAWR
|
PRKC, apoptosis, WT1, regulator |
| chr4_-_103749205 | 0.07 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr20_-_36152914 | 0.06 |
ENST00000397131.1
|
BLCAP
|
bladder cancer associated protein |
| chr1_-_55266926 | 0.06 |
ENST00000371276.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr16_-_74808710 | 0.06 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr9_+_133710453 | 0.06 |
ENST00000318560.5
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr6_-_36355486 | 0.06 |
ENST00000538992.1
|
ETV7
|
ets variant 7 |
| chr2_-_28113217 | 0.06 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr4_-_177116772 | 0.06 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr7_-_78400598 | 0.06 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr22_-_46931191 | 0.06 |
ENST00000454637.1
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr5_-_159739532 | 0.06 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr17_+_37793378 | 0.06 |
ENST00000544210.2
ENST00000581894.1 ENST00000394250.4 ENST00000579479.1 ENST00000577248.1 ENST00000580611.1 |
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr7_-_92855762 | 0.06 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr6_-_42016385 | 0.06 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr19_-_18632861 | 0.06 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr3_-_49170405 | 0.06 |
ENST00000305544.4
ENST00000494831.1 |
LAMB2
|
laminin, beta 2 (laminin S) |
| chr6_-_39197226 | 0.06 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr6_+_31939608 | 0.06 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr2_+_30670209 | 0.06 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr7_+_74379083 | 0.05 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr16_+_83932684 | 0.05 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr4_-_140223670 | 0.05 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr16_-_4465886 | 0.05 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr7_+_99699280 | 0.05 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr19_+_36249044 | 0.05 |
ENST00000444637.2
ENST00000396908.4 ENST00000544099.1 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr16_+_103816 | 0.05 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr15_-_45670924 | 0.05 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr20_+_44044717 | 0.05 |
ENST00000279036.6
ENST00000279035.9 ENST00000372689.5 ENST00000545755.1 ENST00000341555.5 ENST00000535404.1 ENST00000543458.2 ENST00000432270.1 |
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr19_-_40324255 | 0.05 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr1_-_55266865 | 0.05 |
ENST00000371274.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr10_+_104614008 | 0.05 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr2_+_37571717 | 0.05 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr7_+_128784712 | 0.05 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr18_+_77439775 | 0.05 |
ENST00000299543.7
ENST00000075430.7 |
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr14_+_31343747 | 0.05 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr9_-_139258159 | 0.05 |
ENST00000371739.3
|
DNLZ
|
DNL-type zinc finger |
| chr3_-_128294929 | 0.05 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr5_-_132948216 | 0.05 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chr9_+_140172200 | 0.05 |
ENST00000357503.2
|
TOR4A
|
torsin family 4, member A |
| chr17_-_4458616 | 0.05 |
ENST00000381556.2
|
MYBBP1A
|
MYB binding protein (P160) 1a |
| chr16_-_69418553 | 0.05 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr19_-_7939319 | 0.05 |
ENST00000539422.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr6_+_34204642 | 0.05 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_-_11120057 | 0.04 |
ENST00000376957.2
|
SRM
|
spermidine synthase |
| chr11_+_93754513 | 0.04 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr1_+_155051305 | 0.04 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr15_-_75165651 | 0.04 |
ENST00000562363.1
ENST00000564529.1 ENST00000268099.9 |
SCAMP2
|
secretory carrier membrane protein 2 |
| chr9_-_130829588 | 0.04 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr9_-_123555655 | 0.04 |
ENST00000340778.5
ENST00000453291.1 ENST00000608872.1 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr3_-_50383096 | 0.04 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr4_-_76555657 | 0.04 |
ENST00000307465.4
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr11_-_117698787 | 0.04 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr7_-_8302164 | 0.04 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr17_-_29624343 | 0.04 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr2_-_225811747 | 0.04 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr17_+_76164639 | 0.04 |
ENST00000225777.3
ENST00000585591.1 ENST00000589711.1 ENST00000588282.1 ENST00000589168.1 |
SYNGR2
|
synaptogyrin 2 |
| chr12_+_122150646 | 0.04 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr19_+_18794470 | 0.04 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr4_-_103749179 | 0.04 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_10419871 | 0.04 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr8_+_22435762 | 0.04 |
ENST00000456545.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr7_-_2883928 | 0.04 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chrX_+_47863734 | 0.04 |
ENST00000304355.5
|
SPACA5
|
sperm acrosome associated 5 |
| chrX_+_110339439 | 0.04 |
ENST00000372010.1
ENST00000519681.1 ENST00000372007.5 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr14_-_92413353 | 0.04 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr9_-_134406565 | 0.04 |
ENST00000372210.3
ENST00000372211.3 |
UCK1
|
uridine-cytidine kinase 1 |
| chr11_-_108093329 | 0.04 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr4_+_81951957 | 0.04 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr17_-_34890617 | 0.04 |
ENST00000586886.1
ENST00000585719.1 ENST00000585818.1 |
MYO19
|
myosin XIX |
| chr1_+_26737253 | 0.04 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr19_+_50180317 | 0.03 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr12_-_62586543 | 0.03 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr8_-_144512576 | 0.03 |
ENST00000333480.2
|
MAFA
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
| chr17_-_46035187 | 0.03 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr19_-_42721819 | 0.03 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr9_+_33795533 | 0.03 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr13_+_22245522 | 0.03 |
ENST00000382353.5
|
FGF9
|
fibroblast growth factor 9 |
| chr14_+_69726864 | 0.03 |
ENST00000448469.3
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr8_-_93029865 | 0.03 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_-_178937478 | 0.03 |
ENST00000286063.6
|
PDE11A
|
phosphodiesterase 11A |
| chr17_+_76165213 | 0.03 |
ENST00000590201.1
|
SYNGR2
|
synaptogyrin 2 |
| chr14_-_90085458 | 0.03 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr16_+_77224732 | 0.03 |
ENST00000569610.1
ENST00000248248.3 ENST00000567291.1 ENST00000320859.6 ENST00000563612.1 ENST00000563279.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr17_+_37793318 | 0.03 |
ENST00000336308.5
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr14_+_67707826 | 0.03 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr5_-_150467221 | 0.03 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr21_-_39870339 | 0.03 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr7_+_128470431 | 0.03 |
ENST00000325888.8
ENST00000346177.6 |
FLNC
|
filamin C, gamma |
| chr2_-_136288740 | 0.03 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr5_-_150466692 | 0.03 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_+_7107999 | 0.03 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr4_+_5712898 | 0.03 |
ENST00000264956.6
ENST00000382674.2 |
EVC
|
Ellis van Creveld syndrome |
| chr6_+_155537771 | 0.03 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr13_-_41635512 | 0.03 |
ENST00000405737.2
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr12_-_56326402 | 0.03 |
ENST00000547925.1
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr4_-_54457783 | 0.03 |
ENST00000263925.7
ENST00000512247.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr1_-_154928562 | 0.03 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr6_+_7108210 | 0.03 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr19_-_50316517 | 0.03 |
ENST00000313777.4
ENST00000445575.2 |
FUZ
|
fuzzy planar cell polarity protein |
| chr8_-_103668114 | 0.03 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr17_-_1083078 | 0.03 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr17_+_7284365 | 0.03 |
ENST00000311668.2
|
TNK1
|
tyrosine kinase, non-receptor, 1 |
| chr3_-_178984759 | 0.03 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr15_+_90728145 | 0.03 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
Network of associatons between targets according to the STRING database.
First level regulatory network of FIGLA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.0 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.2 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.0 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |