Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for EVX2
Z-value: 0.25
Transcription factors associated with EVX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EVX2
|
ENSG00000174279.4 | even-skipped homeobox 2 |
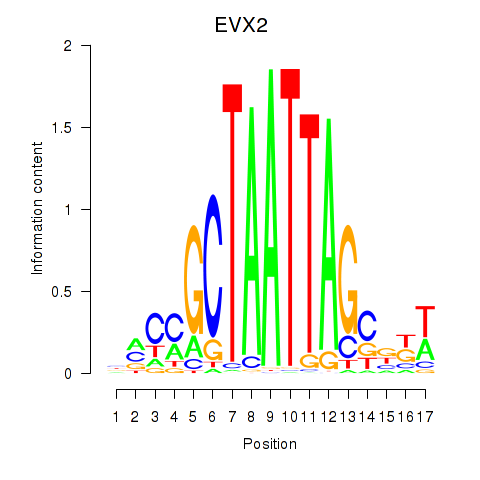
Activity profile of EVX2 motif
Sorted Z-values of EVX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_111718173 | 0.11 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr17_+_37809333 | 0.09 |
ENST00000443521.1
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr15_+_41057818 | 0.07 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr4_-_41884620 | 0.07 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr3_+_111717511 | 0.06 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr13_+_78315528 | 0.06 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_+_111717600 | 0.06 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr3_+_111718036 | 0.06 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr12_+_64798826 | 0.06 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr6_-_62996066 | 0.05 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr15_-_55562582 | 0.04 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55563072 | 0.04 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr20_+_43990576 | 0.04 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr1_+_28261533 | 0.03 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr13_+_78315466 | 0.03 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr19_+_11071546 | 0.03 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr15_-_55562479 | 0.03 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_+_144354644 | 0.02 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr4_+_144312659 | 0.02 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr17_-_40337470 | 0.02 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr17_-_9929581 | 0.01 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr1_-_159832438 | 0.01 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr3_+_115342349 | 0.01 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr20_+_30467600 | 0.01 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr1_+_28261492 | 0.01 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_+_65554493 | 0.01 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr6_-_111927062 | 0.00 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr1_-_151431674 | 0.00 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr12_+_15699286 | 0.00 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
Network of associatons between targets according to the STRING database.
First level regulatory network of EVX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |