Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
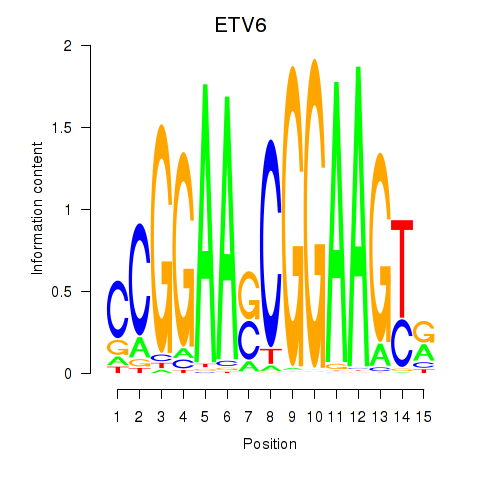
Results for ETV6
Z-value: 1.20
Transcription factors associated with ETV6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV6
|
ENSG00000139083.6 | ETS variant transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV6 | hg19_v2_chr12_+_11802753_11802834 | 0.29 | 5.7e-01 | Click! |
Activity profile of ETV6 motif
Sorted Z-values of ETV6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153066998 | 1.71 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chr15_-_80263506 | 1.14 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr1_-_153044083 | 1.10 |
ENST00000341611.2
|
SPRR2B
|
small proline-rich protein 2B |
| chr17_+_77020325 | 1.06 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020224 | 1.02 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr5_-_150473127 | 0.81 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_+_26156551 | 0.65 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr17_+_77020146 | 0.55 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr16_+_67226019 | 0.53 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding |
| chr1_-_54518865 | 0.51 |
ENST00000371337.3
|
TMEM59
|
transmembrane protein 59 |
| chr11_-_67205538 | 0.50 |
ENST00000326294.3
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr12_+_64846129 | 0.48 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr20_+_2821366 | 0.46 |
ENST00000453689.1
ENST00000417508.1 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr17_+_45908974 | 0.45 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr6_-_11382478 | 0.44 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr2_+_239335636 | 0.42 |
ENST00000409297.1
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr12_-_58329819 | 0.41 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr22_+_38004723 | 0.40 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr19_+_52693259 | 0.40 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr1_-_2323140 | 0.40 |
ENST00000378531.3
ENST00000378529.3 |
MORN1
|
MORN repeat containing 1 |
| chr1_-_246729544 | 0.39 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr6_-_31620095 | 0.39 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr12_+_64845864 | 0.38 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr3_-_16555150 | 0.36 |
ENST00000334133.4
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr3_-_52739670 | 0.36 |
ENST00000497953.1
|
GLT8D1
|
glycosyltransferase 8 domain containing 1 |
| chr19_-_11669960 | 0.36 |
ENST00000589171.1
ENST00000590700.1 ENST00000586683.1 ENST00000593077.1 ENST00000252445.3 |
ELOF1
|
elongation factor 1 homolog (S. cerevisiae) |
| chr5_+_159656437 | 0.35 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr1_-_43855444 | 0.35 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr19_-_9929484 | 0.35 |
ENST00000586651.1
ENST00000586073.1 |
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr17_-_61904420 | 0.34 |
ENST00000580272.1
|
FTSJ3
|
FtsJ homolog 3 (E. coli) |
| chr19_-_54618650 | 0.34 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr6_-_31620403 | 0.34 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr1_+_150266256 | 0.34 |
ENST00000309092.7
ENST00000369084.5 |
MRPS21
|
mitochondrial ribosomal protein S21 |
| chr14_+_100842735 | 0.34 |
ENST00000554998.1
ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25
|
WD repeat domain 25 |
| chr16_+_69373471 | 0.33 |
ENST00000569637.2
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr2_+_38152462 | 0.33 |
ENST00000354545.2
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr20_+_58713514 | 0.32 |
ENST00000432910.1
|
RP5-1043L13.1
|
RP5-1043L13.1 |
| chr14_-_24711865 | 0.31 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr22_+_38004832 | 0.31 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr10_+_44101850 | 0.31 |
ENST00000361807.3
ENST00000374437.2 ENST00000430885.1 ENST00000374435.3 |
ZNF485
|
zinc finger protein 485 |
| chr9_-_100684769 | 0.30 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chr19_-_59066452 | 0.30 |
ENST00000312547.2
|
CHMP2A
|
charged multivesicular body protein 2A |
| chr22_+_38005033 | 0.30 |
ENST00000447515.1
ENST00000406772.1 ENST00000431745.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr6_-_31620455 | 0.30 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr6_-_43496605 | 0.29 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr19_+_49497121 | 0.29 |
ENST00000413176.2
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr20_-_34542548 | 0.29 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr1_+_15736359 | 0.29 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr19_+_49496782 | 0.29 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr19_-_9938480 | 0.28 |
ENST00000585379.1
|
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr19_-_10230562 | 0.28 |
ENST00000587146.1
ENST00000588709.1 ENST00000253108.4 |
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr7_+_155090271 | 0.28 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr12_-_123459105 | 0.28 |
ENST00000543935.1
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr6_-_31620149 | 0.27 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr16_-_420338 | 0.27 |
ENST00000450882.1
ENST00000441883.1 ENST00000447696.1 ENST00000389675.2 |
MRPL28
|
mitochondrial ribosomal protein L28 |
| chr16_+_30406423 | 0.27 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr6_-_31619892 | 0.26 |
ENST00000454165.1
ENST00000428326.1 ENST00000452994.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr7_-_91509972 | 0.26 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr16_+_69373323 | 0.26 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr14_-_24711470 | 0.25 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr2_-_27632390 | 0.25 |
ENST00000350803.4
ENST00000344034.4 |
PPM1G
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr9_+_97021572 | 0.25 |
ENST00000395395.2
ENST00000340911.4 |
ZNF169
|
zinc finger protein 169 |
| chr1_+_24069952 | 0.25 |
ENST00000609199.1
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr6_-_42946888 | 0.25 |
ENST00000244546.4
|
PEX6
|
peroxisomal biogenesis factor 6 |
| chr2_+_172543919 | 0.24 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr16_-_54963026 | 0.24 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr19_+_49496705 | 0.24 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr7_-_99756293 | 0.24 |
ENST00000316937.3
ENST00000456769.1 |
C7orf43
|
chromosome 7 open reading frame 43 |
| chr14_-_24711764 | 0.24 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr19_-_19302931 | 0.24 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr6_-_91296737 | 0.24 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr14_-_24711806 | 0.24 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_-_10362725 | 0.24 |
ENST00000397109.3
ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13
|
SEC13 homolog (S. cerevisiae) |
| chr19_-_56826157 | 0.23 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr1_-_1208851 | 0.23 |
ENST00000488418.1
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr19_+_49375649 | 0.23 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr5_+_73109339 | 0.23 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr7_+_44240520 | 0.23 |
ENST00000496112.1
ENST00000223369.2 |
YKT6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr15_+_91427726 | 0.23 |
ENST00000452243.1
|
FES
|
feline sarcoma oncogene |
| chr1_+_32687971 | 0.23 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr12_-_120907374 | 0.22 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr16_-_74330612 | 0.22 |
ENST00000569389.1
ENST00000562888.1 |
AC009120.4
|
AC009120.4 |
| chr15_+_74908228 | 0.22 |
ENST00000566126.1
|
CLK3
|
CDC-like kinase 3 |
| chr3_-_16554881 | 0.22 |
ENST00000451036.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr10_-_99161033 | 0.22 |
ENST00000315563.6
ENST00000370992.4 ENST00000414986.1 |
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr1_+_43855560 | 0.22 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr19_-_3971050 | 0.22 |
ENST00000545797.2
ENST00000596311.1 |
DAPK3
|
death-associated protein kinase 3 |
| chr1_+_24069642 | 0.22 |
ENST00000418390.2
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr14_-_21852119 | 0.21 |
ENST00000555943.1
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae) |
| chr15_-_43559055 | 0.21 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr17_+_79935464 | 0.21 |
ENST00000581647.1
ENST00000580534.1 ENST00000579684.1 |
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr14_-_45431091 | 0.20 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr19_-_45737469 | 0.20 |
ENST00000413988.1
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr19_-_16682987 | 0.20 |
ENST00000431408.1
ENST00000436553.2 ENST00000595753.1 |
SLC35E1
|
solute carrier family 35, member E1 |
| chr2_-_96971232 | 0.20 |
ENST00000323853.5
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5) |
| chr16_-_67694129 | 0.20 |
ENST00000602320.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr2_+_113885138 | 0.20 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr19_+_56186606 | 0.20 |
ENST00000085079.7
|
EPN1
|
epsin 1 |
| chr17_-_48450534 | 0.20 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr2_+_239335449 | 0.20 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr9_-_139268068 | 0.20 |
ENST00000371734.3
ENST00000371732.5 ENST00000315908.7 |
CARD9
|
caspase recruitment domain family, member 9 |
| chr22_+_40742497 | 0.20 |
ENST00000216194.7
|
ADSL
|
adenylosuccinate lyase |
| chr22_-_42084863 | 0.19 |
ENST00000401959.1
ENST00000355257.3 |
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr6_-_33256664 | 0.19 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr2_+_172543967 | 0.19 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr12_-_120907459 | 0.19 |
ENST00000229390.3
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr11_+_65770227 | 0.19 |
ENST00000527348.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr19_-_10514184 | 0.19 |
ENST00000589629.1
ENST00000222005.2 |
CDC37
|
cell division cycle 37 |
| chr21_-_46340770 | 0.19 |
ENST00000397854.3
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr8_+_104427581 | 0.19 |
ENST00000521716.1
ENST00000521971.1 ENST00000519682.1 |
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr3_+_111805262 | 0.19 |
ENST00000484828.1
|
C3orf52
|
chromosome 3 open reading frame 52 |
| chr6_-_91296602 | 0.19 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr5_+_36152091 | 0.19 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr6_+_30294612 | 0.19 |
ENST00000440271.1
ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39
|
tripartite motif containing 39 |
| chr19_-_54619006 | 0.19 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr14_+_24701870 | 0.19 |
ENST00000561035.1
ENST00000559409.1 ENST00000558865.1 ENST00000558279.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr15_+_75074915 | 0.19 |
ENST00000567123.1
ENST00000569462.1 |
CSK
|
c-src tyrosine kinase |
| chr17_+_18218587 | 0.19 |
ENST00000406438.3
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr4_-_185269050 | 0.19 |
ENST00000511465.1
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr2_+_99771527 | 0.19 |
ENST00000415142.1
ENST00000436234.1 |
LIPT1
|
lipoyltransferase 1 |
| chr6_-_31619697 | 0.19 |
ENST00000434444.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr11_+_70244510 | 0.19 |
ENST00000346329.3
ENST00000301843.8 ENST00000376561.3 |
CTTN
|
cortactin |
| chr15_+_91427691 | 0.18 |
ENST00000559355.1
ENST00000394302.1 |
FES
|
feline sarcoma oncogene |
| chr19_-_13044494 | 0.18 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr6_-_656963 | 0.18 |
ENST00000380907.2
|
HUS1B
|
HUS1 checkpoint homolog b (S. pombe) |
| chr6_-_31697563 | 0.18 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr4_-_10118469 | 0.18 |
ENST00000499869.2
|
WDR1
|
WD repeat domain 1 |
| chr11_-_116658758 | 0.18 |
ENST00000227322.3
|
ZNF259
|
zinc finger protein 259 |
| chr19_+_56186557 | 0.18 |
ENST00000270460.6
|
EPN1
|
epsin 1 |
| chr3_-_169482840 | 0.18 |
ENST00000602385.1
|
TERC
|
telomerase RNA component |
| chr19_+_50919056 | 0.18 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr1_-_55266865 | 0.18 |
ENST00000371274.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr17_-_55927370 | 0.18 |
ENST00000578444.1
ENST00000313608.8 ENST00000579380.1 |
MRPS23
|
mitochondrial ribosomal protein S23 |
| chr6_+_30295036 | 0.17 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr2_+_96931834 | 0.17 |
ENST00000488633.1
|
CIAO1
|
cytosolic iron-sulfur protein assembly 1 |
| chr11_+_7041677 | 0.17 |
ENST00000299481.4
|
NLRP14
|
NLR family, pyrin domain containing 14 |
| chr1_-_43855479 | 0.17 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr17_-_26989136 | 0.17 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr5_+_147763498 | 0.17 |
ENST00000340253.5
|
FBXO38
|
F-box protein 38 |
| chr17_+_46048497 | 0.17 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr2_-_95831158 | 0.17 |
ENST00000447814.1
|
ZNF514
|
zinc finger protein 514 |
| chr21_-_46340884 | 0.17 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr17_+_46048471 | 0.17 |
ENST00000578018.1
ENST00000579175.1 |
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr11_+_47600562 | 0.17 |
ENST00000263774.4
ENST00000529276.1 ENST00000528192.1 ENST00000530295.1 ENST00000534208.1 ENST00000534716.2 |
NDUFS3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| chr11_+_68671310 | 0.17 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr12_+_27863706 | 0.17 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr6_+_43603552 | 0.17 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr11_+_64851666 | 0.17 |
ENST00000525509.1
ENST00000294258.3 ENST00000526334.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr6_-_125623046 | 0.17 |
ENST00000608295.1
ENST00000398153.2 ENST00000608284.1 ENST00000368377.4 |
HDDC2
|
HD domain containing 2 |
| chr14_+_105864847 | 0.16 |
ENST00000451127.2
|
TEX22
|
testis expressed 22 |
| chr8_-_104427313 | 0.16 |
ENST00000297578.4
|
SLC25A32
|
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr15_+_75074410 | 0.16 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr17_-_76836729 | 0.16 |
ENST00000587783.1
ENST00000542802.3 ENST00000586531.1 ENST00000589424.1 ENST00000590546.2 |
USP36
|
ubiquitin specific peptidase 36 |
| chr16_-_67693846 | 0.16 |
ENST00000602850.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr19_-_6393216 | 0.16 |
ENST00000595047.1
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa |
| chr11_+_327171 | 0.16 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr5_+_34839260 | 0.16 |
ENST00000505624.1
|
TTC23L
|
tetratricopeptide repeat domain 23-like |
| chr11_-_6704513 | 0.16 |
ENST00000532203.1
ENST00000288937.6 |
MRPL17
|
mitochondrial ribosomal protein L17 |
| chr8_-_145980808 | 0.16 |
ENST00000525191.1
|
ZNF251
|
zinc finger protein 251 |
| chr4_-_10118573 | 0.16 |
ENST00000382452.2
ENST00000382451.2 |
WDR1
|
WD repeat domain 1 |
| chr17_-_2239729 | 0.16 |
ENST00000576112.2
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr1_+_156698743 | 0.16 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr17_+_30813576 | 0.16 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr10_+_26986582 | 0.16 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr17_+_4843303 | 0.16 |
ENST00000571816.1
|
RNF167
|
ring finger protein 167 |
| chr17_-_61777459 | 0.16 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr16_-_15736953 | 0.16 |
ENST00000548025.1
ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430
|
KIAA0430 |
| chr21_+_46654249 | 0.16 |
ENST00000584169.1
ENST00000328344.2 |
LINC00334
|
long intergenic non-protein coding RNA 334 |
| chr6_+_31633833 | 0.16 |
ENST00000375882.2
ENST00000375880.2 |
CSNK2B
CSNK2B-LY6G5B-1181
|
casein kinase 2, beta polypeptide Uncharacterized protein |
| chr11_-_18548426 | 0.15 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chr10_+_75504105 | 0.15 |
ENST00000535742.1
ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C
|
SEC24 family member C |
| chr6_-_31697255 | 0.15 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_-_116658695 | 0.15 |
ENST00000429220.1
ENST00000444935.1 |
ZNF259
|
zinc finger protein 259 |
| chr19_+_1275917 | 0.15 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr1_-_202858227 | 0.15 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chr11_-_65625014 | 0.15 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr17_-_57784755 | 0.15 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr1_-_38325256 | 0.15 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr17_-_15587602 | 0.15 |
ENST00000416464.2
ENST00000578237.1 ENST00000581200.1 |
TRIM16
|
tripartite motif containing 16 |
| chr7_+_44084262 | 0.15 |
ENST00000456905.1
ENST00000440166.1 ENST00000452943.1 ENST00000468694.1 ENST00000494774.1 ENST00000490734.2 |
DBNL
|
drebrin-like |
| chr8_-_124054484 | 0.15 |
ENST00000419562.2
|
DERL1
|
derlin 1 |
| chr6_+_30524663 | 0.15 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr18_-_45456930 | 0.15 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr4_-_5710257 | 0.15 |
ENST00000344938.1
ENST00000344408.5 |
EVC2
|
Ellis van Creveld syndrome 2 |
| chr3_+_45635661 | 0.15 |
ENST00000440097.1
|
LIMD1
|
LIM domains containing 1 |
| chr2_+_241544834 | 0.15 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr1_+_84944926 | 0.15 |
ENST00000370656.1
ENST00000370654.5 |
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr1_+_154193643 | 0.15 |
ENST00000456325.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr14_-_100842588 | 0.15 |
ENST00000556645.1
ENST00000556209.1 ENST00000556504.1 ENST00000556435.1 ENST00000554772.1 ENST00000553581.1 ENST00000553769.2 ENST00000554605.1 ENST00000557722.1 ENST00000553413.1 ENST00000553524.1 ENST00000358655.4 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr19_+_56166360 | 0.15 |
ENST00000308924.4
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr1_-_19811962 | 0.15 |
ENST00000375142.1
ENST00000264202.6 |
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr6_-_31619742 | 0.15 |
ENST00000433828.1
ENST00000456286.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr19_+_11546440 | 0.15 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr6_+_57182400 | 0.15 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr6_-_31697977 | 0.15 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr19_-_1876156 | 0.15 |
ENST00000565797.1
|
CTB-31O20.2
|
CTB-31O20.2 |
| chr19_-_10230540 | 0.15 |
ENST00000589454.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr2_+_201981119 | 0.14 |
ENST00000395148.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr10_+_102729249 | 0.14 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr1_-_43638168 | 0.14 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.9 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.2 | 0.5 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 0.8 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 0.8 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.4 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 1.0 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.5 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.5 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 1.0 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.3 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.2 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.2 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 0.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.2 | GO:2000397 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.4 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 3.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.2 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0052363 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.3 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.6 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.4 | GO:0007252 | activation of NF-kappaB-inducing kinase activity(GO:0007250) I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.2 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation(GO:0060051) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.2 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.8 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.2 | 1.9 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0042565 | nuclear RNA export factor complex(GO:0042272) RNA nuclear export complex(GO:0042565) |
| 0.1 | 0.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 0.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 2.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.5 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.7 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.2 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 1.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 2.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.0 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 1.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |