Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
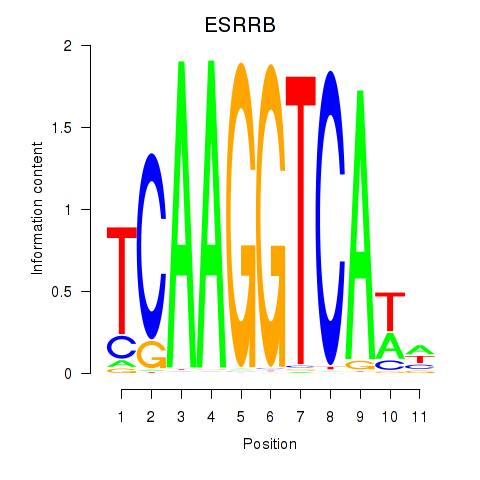
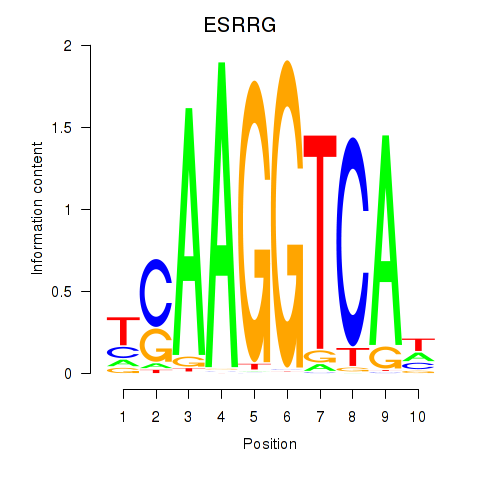
Results for ESRRB_ESRRG
Z-value: 1.21
Transcription factors associated with ESRRB_ESRRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESRRB
|
ENSG00000119715.10 | estrogen related receptor beta |
|
ESRRG
|
ENSG00000196482.12 | estrogen related receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESRRB | hg19_v2_chr14_+_76776957_76777061 | -0.70 | 1.2e-01 | Click! |
| ESRRG | hg19_v2_chr1_-_216896780_216896800 | 0.50 | 3.1e-01 | Click! |
Activity profile of ESRRB_ESRRG motif
Sorted Z-values of ESRRB_ESRRG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_74864386 | 1.38 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr6_-_72129806 | 1.16 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr16_+_58535372 | 1.04 |
ENST00000566656.1
ENST00000566618.1 |
NDRG4
|
NDRG family member 4 |
| chr19_+_42824511 | 1.04 |
ENST00000601644.1
|
TMEM145
|
transmembrane protein 145 |
| chrX_+_109602039 | 0.69 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr15_+_67814008 | 0.66 |
ENST00000557807.1
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr14_+_21492331 | 0.62 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr14_-_31889737 | 0.62 |
ENST00000382464.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr1_+_16083098 | 0.62 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr2_+_228678550 | 0.58 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr19_+_589893 | 0.56 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr6_+_31916733 | 0.51 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr17_+_76037081 | 0.49 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr1_-_153433120 | 0.48 |
ENST00000368723.3
|
S100A7
|
S100 calcium binding protein A7 |
| chr17_+_73455788 | 0.46 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr7_+_76107444 | 0.45 |
ENST00000435861.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr15_+_67813406 | 0.44 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr17_-_79792909 | 0.44 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr8_-_100905925 | 0.43 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr10_+_112257596 | 0.42 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr11_-_132813566 | 0.41 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr6_+_43968306 | 0.40 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr11_-_68611721 | 0.40 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr7_-_127671674 | 0.40 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr18_-_43678241 | 0.39 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr11_+_10472223 | 0.39 |
ENST00000396554.3
ENST00000524866.1 |
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr8_-_100905850 | 0.38 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr16_+_28874345 | 0.37 |
ENST00000566209.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr17_-_65235916 | 0.36 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr5_-_150473127 | 0.36 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr15_-_91475706 | 0.36 |
ENST00000561036.1
|
HDDC3
|
HD domain containing 3 |
| chr1_+_161172143 | 0.36 |
ENST00000476409.2
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) |
| chr2_+_228190066 | 0.35 |
ENST00000436237.1
ENST00000443428.2 ENST00000418961.1 |
MFF
|
mitochondrial fission factor |
| chr13_-_50020554 | 0.34 |
ENST00000400396.1
|
AL136218.1
|
Sarcoma antigen NY-SAR-79; Uncharacterized protein |
| chr8_-_100905363 | 0.33 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr1_-_207119738 | 0.31 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr21_-_46330545 | 0.31 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr19_+_7011509 | 0.31 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr3_-_52312337 | 0.30 |
ENST00000469000.1
|
WDR82
|
WD repeat domain 82 |
| chrX_+_48620147 | 0.30 |
ENST00000303227.6
|
GLOD5
|
glyoxalase domain containing 5 |
| chr19_+_46850320 | 0.30 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr15_+_59730348 | 0.29 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr14_-_91282821 | 0.29 |
ENST00000553948.1
|
TTC7B
|
tetratricopeptide repeat domain 7B |
| chr1_+_169077133 | 0.29 |
ENST00000494797.1
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr14_+_51026844 | 0.28 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr14_-_103987679 | 0.28 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr7_-_68895857 | 0.27 |
ENST00000421513.1
|
RP11-3P22.2
|
RP11-3P22.2 |
| chrX_-_154255143 | 0.27 |
ENST00000453950.1
ENST00000423959.1 |
F8
|
coagulation factor VIII, procoagulant component |
| chr16_-_4588391 | 0.26 |
ENST00000586728.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr8_-_131028782 | 0.26 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr3_+_102153859 | 0.26 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr3_-_179322436 | 0.25 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr11_+_118272328 | 0.25 |
ENST00000524422.1
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr1_+_29213678 | 0.25 |
ENST00000347529.3
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr1_-_23504176 | 0.25 |
ENST00000302291.4
|
LUZP1
|
leucine zipper protein 1 |
| chr12_+_58176525 | 0.24 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr22_+_37959647 | 0.24 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr5_+_71403280 | 0.24 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr19_-_7293942 | 0.24 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr16_-_3767551 | 0.24 |
ENST00000246957.5
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr17_+_900342 | 0.24 |
ENST00000327158.4
|
TIMM22
|
translocase of inner mitochondrial membrane 22 homolog (yeast) |
| chr11_+_65770227 | 0.24 |
ENST00000527348.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr11_+_67798114 | 0.23 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr3_+_10290596 | 0.23 |
ENST00000448281.2
|
TATDN2
|
TatD DNase domain containing 2 |
| chr10_-_76995769 | 0.23 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr2_+_198365095 | 0.23 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr8_+_110346546 | 0.23 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr8_+_145149930 | 0.23 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr12_-_120554534 | 0.23 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr17_-_1463095 | 0.23 |
ENST00000575895.1
ENST00000573056.1 |
PITPNA
|
phosphatidylinositol transfer protein, alpha |
| chr5_+_80529104 | 0.22 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr16_-_3767506 | 0.22 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr18_-_56985776 | 0.22 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr17_+_46970134 | 0.22 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr19_+_38308119 | 0.22 |
ENST00000592103.1
|
CTD-2554C21.2
|
CTD-2554C21.2 |
| chr6_-_13290684 | 0.22 |
ENST00000606393.1
|
RP1-257A7.5
|
RP1-257A7.5 |
| chr17_+_79670386 | 0.22 |
ENST00000333676.3
ENST00000571730.1 ENST00000541223.1 |
MRPL12
SLC25A10
SLC25A10
|
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr18_-_12377001 | 0.22 |
ENST00000590811.1
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr19_+_46850251 | 0.22 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr14_-_58893832 | 0.21 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_-_91475747 | 0.21 |
ENST00000330334.3
ENST00000559898.1 ENST00000394272.3 |
HDDC3
|
HD domain containing 3 |
| chr20_+_30458431 | 0.21 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr2_+_238875597 | 0.21 |
ENST00000272930.4
ENST00000448502.1 ENST00000416292.1 ENST00000409633.1 ENST00000414443.1 ENST00000409953.1 ENST00000409332.1 |
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr1_+_16083123 | 0.21 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr4_+_76439649 | 0.20 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr8_-_90769422 | 0.20 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr12_-_57039739 | 0.20 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr16_+_691792 | 0.20 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr1_+_169077172 | 0.20 |
ENST00000499679.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr3_-_10028366 | 0.20 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr22_+_30821784 | 0.20 |
ENST00000407550.3
|
MTFP1
|
mitochondrial fission process 1 |
| chr4_-_140216948 | 0.19 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr6_+_30749649 | 0.19 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr20_+_31755934 | 0.19 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr1_+_155179012 | 0.19 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr15_+_78441663 | 0.19 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr1_-_33502528 | 0.19 |
ENST00000354858.6
|
AK2
|
adenylate kinase 2 |
| chr11_+_3968573 | 0.19 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr8_-_102275131 | 0.19 |
ENST00000523121.1
|
KB-1410C5.2
|
KB-1410C5.2 |
| chr2_+_172950227 | 0.19 |
ENST00000341900.6
|
DLX1
|
distal-less homeobox 1 |
| chr9_-_130889990 | 0.19 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr19_+_57742369 | 0.19 |
ENST00000415300.2
ENST00000448930.1 |
AURKC
|
aurora kinase C |
| chr15_+_67458861 | 0.19 |
ENST00000558428.1
ENST00000558827.1 |
SMAD3
|
SMAD family member 3 |
| chr1_-_17380630 | 0.18 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr13_-_111214015 | 0.18 |
ENST00000267328.3
|
RAB20
|
RAB20, member RAS oncogene family |
| chr3_+_184037466 | 0.18 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr19_-_18314799 | 0.18 |
ENST00000481914.2
|
RAB3A
|
RAB3A, member RAS oncogene family |
| chr16_+_777118 | 0.18 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr9_+_134001455 | 0.18 |
ENST00000531584.1
|
NUP214
|
nucleoporin 214kDa |
| chr14_-_75536182 | 0.18 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr11_+_57480046 | 0.18 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr15_-_41694640 | 0.18 |
ENST00000558719.1
ENST00000260361.4 ENST00000560978.1 |
NDUFAF1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr2_-_176046391 | 0.17 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr1_+_161195835 | 0.17 |
ENST00000545897.1
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr3_+_184038073 | 0.17 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_165600436 | 0.17 |
ENST00000367888.4
ENST00000367885.1 ENST00000367884.2 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr3_+_128968437 | 0.17 |
ENST00000314797.6
|
COPG1
|
coatomer protein complex, subunit gamma 1 |
| chr2_+_145425573 | 0.17 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_-_132812987 | 0.17 |
ENST00000541867.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr7_+_44836276 | 0.17 |
ENST00000451562.1
ENST00000468812.1 ENST00000489459.1 ENST00000355968.6 |
PPIA
|
peptidylprolyl isomerase A (cyclophilin A) |
| chr7_-_139876734 | 0.17 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr19_+_35521572 | 0.17 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr22_+_30792980 | 0.17 |
ENST00000403484.1
ENST00000405717.3 ENST00000402592.3 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr19_-_55574538 | 0.17 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr2_-_86422095 | 0.17 |
ENST00000254636.5
|
IMMT
|
inner membrane protein, mitochondrial |
| chr1_-_33502441 | 0.17 |
ENST00000548033.1
ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2
|
adenylate kinase 2 |
| chr12_+_109915422 | 0.17 |
ENST00000280774.5
ENST00000536398.1 ENST00000539599.1 ENST00000342494.3 ENST00000340074.5 ENST00000540230.1 ENST00000537063.1 |
UBE3B
|
ubiquitin protein ligase E3B |
| chr9_+_112852477 | 0.17 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr8_+_119294456 | 0.17 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chrX_-_154033661 | 0.16 |
ENST00000393531.1
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr1_-_159825137 | 0.16 |
ENST00000368102.1
|
C1orf204
|
chromosome 1 open reading frame 204 |
| chr13_+_22245522 | 0.16 |
ENST00000382353.5
|
FGF9
|
fibroblast growth factor 9 |
| chr12_-_30907822 | 0.16 |
ENST00000540436.1
|
CAPRIN2
|
caprin family member 2 |
| chr3_+_179065474 | 0.16 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr8_+_86376081 | 0.16 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr15_-_42783303 | 0.16 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr16_-_74734672 | 0.15 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr10_-_123734683 | 0.15 |
ENST00000369017.5
ENST00000369023.3 |
NSMCE4A
|
non-SMC element 4 homolog A (S. cerevisiae) |
| chr1_+_161195781 | 0.15 |
ENST00000367988.3
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr2_-_31030277 | 0.15 |
ENST00000534090.2
ENST00000295055.8 |
CAPN13
|
calpain 13 |
| chr17_-_46799872 | 0.15 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr17_-_49198216 | 0.15 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr4_+_2470664 | 0.15 |
ENST00000314289.8
ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4
|
ring finger protein 4 |
| chrX_-_46187069 | 0.15 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr16_+_85833274 | 0.15 |
ENST00000568794.1
ENST00000253452.2 ENST00000562336.1 ENST00000561569.1 ENST00000566405.1 |
COX4I1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr2_-_220119280 | 0.15 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chr8_+_101349823 | 0.15 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr15_+_45021183 | 0.15 |
ENST00000559390.1
|
TRIM69
|
tripartite motif containing 69 |
| chr5_-_150537279 | 0.15 |
ENST00000517486.1
ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6
|
annexin A6 |
| chr11_-_34379546 | 0.15 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr14_-_21492113 | 0.15 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr1_-_154164534 | 0.15 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chrX_-_46618490 | 0.14 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr20_+_58179582 | 0.14 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr4_-_76957214 | 0.14 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr2_+_145425534 | 0.14 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr4_-_108204904 | 0.14 |
ENST00000510463.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr9_-_130637244 | 0.14 |
ENST00000373156.1
|
AK1
|
adenylate kinase 1 |
| chr7_+_129007964 | 0.14 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr3_-_16306432 | 0.14 |
ENST00000383775.4
ENST00000488423.1 |
DPH3
|
diphthamide biosynthesis 3 |
| chr14_-_24729251 | 0.14 |
ENST00000559136.1
|
TGM1
|
transglutaminase 1 |
| chr16_+_2588012 | 0.13 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr1_+_169075554 | 0.13 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr17_+_55334364 | 0.13 |
ENST00000322684.3
ENST00000579590.1 |
MSI2
|
musashi RNA-binding protein 2 |
| chr3_+_10068095 | 0.13 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr11_+_77899842 | 0.13 |
ENST00000530267.1
|
USP35
|
ubiquitin specific peptidase 35 |
| chr16_-_74734742 | 0.13 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr5_+_177540444 | 0.13 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chr5_+_34757309 | 0.13 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr2_-_128432639 | 0.13 |
ENST00000545738.2
ENST00000355119.4 ENST00000409808.2 |
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr3_+_16306691 | 0.13 |
ENST00000285083.5
ENST00000605932.1 ENST00000435829.2 |
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr19_+_57742431 | 0.13 |
ENST00000302804.7
|
AURKC
|
aurora kinase C |
| chrX_-_154689596 | 0.13 |
ENST00000369444.2
|
H2AFB3
|
H2A histone family, member B3 |
| chr1_+_44444865 | 0.13 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr12_+_51632666 | 0.13 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr17_-_76778339 | 0.13 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr14_-_105262055 | 0.13 |
ENST00000349310.3
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr5_+_38445641 | 0.13 |
ENST00000397210.3
ENST00000506135.1 ENST00000508131.1 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
| chr14_-_105262016 | 0.12 |
ENST00000407796.2
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr10_+_60028818 | 0.12 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr15_-_83621435 | 0.12 |
ENST00000450735.2
ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2
|
homer homolog 2 (Drosophila) |
| chr16_+_718147 | 0.12 |
ENST00000561929.1
|
RHOT2
|
ras homolog family member T2 |
| chr11_+_111957497 | 0.12 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr10_+_105156364 | 0.12 |
ENST00000369797.3
|
PDCD11
|
programmed cell death 11 |
| chr3_-_167452262 | 0.12 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr15_+_36887069 | 0.12 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr16_+_2587965 | 0.12 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr6_+_107349392 | 0.12 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr17_-_67323385 | 0.12 |
ENST00000588665.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr1_+_201924619 | 0.12 |
ENST00000367287.4
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr10_+_81107271 | 0.12 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr22_+_30821732 | 0.12 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr11_+_77899920 | 0.12 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr3_-_49377499 | 0.12 |
ENST00000265560.4
|
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr19_+_38307999 | 0.12 |
ENST00000589653.1
ENST00000590433.1 |
CTD-2554C21.2
|
CTD-2554C21.2 |
| chrX_-_154033686 | 0.12 |
ENST00000453245.1
ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr15_+_43885799 | 0.11 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr3_+_113465866 | 0.11 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr8_-_125869733 | 0.11 |
ENST00000533496.1
|
RP11-1082L8.3
|
RP11-1082L8.3 |
| chr5_+_73109339 | 0.11 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ESRRB_ESRRG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 1.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.5 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.6 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.2 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.2 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.2 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 1.9 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.1 | 0.3 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 1.9 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.2 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.2 | GO:0042938 | positive regulation of cellular pH reduction(GO:0032849) dipeptide transport(GO:0042938) |
| 0.1 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.6 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.2 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.0 | 1.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.0 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.0 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) maintenance of synapse structure(GO:0099558) presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:1902824 | cleavage furrow ingression(GO:0036090) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.2 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.4 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.0 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.6 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0035404 | histone-serine phosphorylation(GO:0035404) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.3 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.0 | GO:0060262 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.1 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.0 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.2 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.0 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.0 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.6 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.4 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.4 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 1.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.2 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.3 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.2 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | ST GAQ PATHWAY | G alpha q Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |