Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
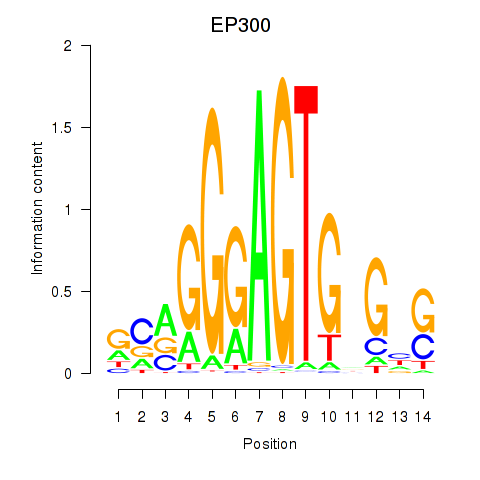
Results for EP300
Z-value: 0.67
Transcription factors associated with EP300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EP300
|
ENSG00000100393.9 | E1A binding protein p300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EP300 | hg19_v2_chr22_+_41487711_41487798 | -0.62 | 1.9e-01 | Click! |
Activity profile of EP300 motif
Sorted Z-values of EP300 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_102188272 | 0.60 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr11_+_102188224 | 0.56 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr12_+_72058130 | 0.55 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr6_-_30685214 | 0.44 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr10_+_81892477 | 0.35 |
ENST00000372263.3
|
PLAC9
|
placenta-specific 9 |
| chr17_-_14140166 | 0.32 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr4_-_74853897 | 0.30 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr18_+_5238549 | 0.27 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr11_-_114271139 | 0.27 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr12_-_122241812 | 0.26 |
ENST00000538335.1
|
AC084018.1
|
AC084018.1 |
| chr19_-_1174226 | 0.25 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr1_+_206643806 | 0.24 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr19_+_49617581 | 0.23 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr1_-_157108266 | 0.23 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr1_+_206643787 | 0.22 |
ENST00000367120.3
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr15_-_74726283 | 0.22 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr10_+_102729249 | 0.21 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr3_-_130745403 | 0.20 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr14_-_69445793 | 0.19 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr6_-_30684898 | 0.19 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr20_+_19738792 | 0.18 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr1_-_209957882 | 0.18 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr16_+_69373471 | 0.18 |
ENST00000569637.2
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr6_+_126240442 | 0.18 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr16_+_69373323 | 0.17 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr8_-_33424636 | 0.17 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr5_+_443280 | 0.17 |
ENST00000508022.1
|
EXOC3
|
exocyst complex component 3 |
| chr20_+_58630972 | 0.17 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr11_-_57194550 | 0.17 |
ENST00000528187.1
ENST00000524863.1 ENST00000533051.1 ENST00000529494.1 ENST00000395124.1 ENST00000533524.1 ENST00000533245.1 ENST00000530316.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr14_-_69445968 | 0.16 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr10_+_105127704 | 0.16 |
ENST00000369839.3
ENST00000351396.4 |
TAF5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
| chr11_-_57194418 | 0.16 |
ENST00000395123.2
ENST00000530005.1 ENST00000532278.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr17_+_7255208 | 0.16 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr1_+_11866270 | 0.15 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr5_+_130506629 | 0.15 |
ENST00000510516.1
ENST00000507584.1 |
LYRM7
|
LYR motif containing 7 |
| chr7_-_139756791 | 0.15 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr12_-_49525175 | 0.14 |
ENST00000336023.5
ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B
|
tubulin, alpha 1b |
| chr19_-_6110457 | 0.14 |
ENST00000586302.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr3_+_171758344 | 0.14 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr14_-_24711865 | 0.14 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr7_-_944631 | 0.14 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr10_+_30723533 | 0.14 |
ENST00000413724.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr2_+_232573208 | 0.14 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr1_+_20915409 | 0.13 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr1_+_11866207 | 0.13 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr14_-_38064198 | 0.13 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr14_+_77582905 | 0.13 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr17_-_19265855 | 0.13 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr1_-_95007193 | 0.13 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr11_+_129685707 | 0.13 |
ENST00000281441.3
|
TMEM45B
|
transmembrane protein 45B |
| chr1_+_38478432 | 0.12 |
ENST00000537711.1
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr13_-_36871886 | 0.12 |
ENST00000491049.2
ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169
SOHLH2
CCDC169-SOHLH2
|
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr1_-_40237020 | 0.12 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr9_-_133814455 | 0.12 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr6_-_149969829 | 0.12 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr17_-_19265982 | 0.12 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr14_-_69446034 | 0.12 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr3_-_158450475 | 0.12 |
ENST00000237696.5
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr14_-_24711470 | 0.12 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr22_+_46731596 | 0.12 |
ENST00000381019.3
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr1_-_1590418 | 0.12 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr12_+_52463751 | 0.12 |
ENST00000336854.4
ENST00000550604.1 ENST00000553049.1 ENST00000548915.1 |
C12orf44
|
chromosome 12 open reading frame 44 |
| chrX_+_23685653 | 0.12 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr1_+_38478378 | 0.11 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr3_+_19988736 | 0.11 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr1_-_54518865 | 0.11 |
ENST00000371337.3
|
TMEM59
|
transmembrane protein 59 |
| chr14_-_24711806 | 0.11 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr6_+_43738444 | 0.11 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr4_+_56814968 | 0.11 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr7_+_90032821 | 0.11 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr20_-_30311703 | 0.11 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr14_-_24711764 | 0.11 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr1_+_157963391 | 0.11 |
ENST00000359209.6
ENST00000416935.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr7_-_5569588 | 0.11 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr1_+_3541543 | 0.11 |
ENST00000378344.2
ENST00000344579.5 |
TPRG1L
|
tumor protein p63 regulated 1-like |
| chr2_+_232573222 | 0.11 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr12_-_124118151 | 0.11 |
ENST00000534960.1
|
EIF2B1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa |
| chr5_+_89770696 | 0.10 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr1_-_155658260 | 0.10 |
ENST00000368339.5
ENST00000405763.3 ENST00000368340.5 ENST00000454523.1 ENST00000443231.1 ENST00000347088.5 ENST00000361831.5 ENST00000355499.4 |
YY1AP1
|
YY1 associated protein 1 |
| chr1_+_155657737 | 0.10 |
ENST00000471642.2
ENST00000471214.1 |
DAP3
|
death associated protein 3 |
| chr11_+_126081662 | 0.10 |
ENST00000528985.1
ENST00000529731.1 ENST00000360194.4 ENST00000530043.1 |
FAM118B
|
family with sequence similarity 118, member B |
| chr16_-_66968055 | 0.10 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr21_+_30671189 | 0.10 |
ENST00000286800.3
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr17_+_21188012 | 0.10 |
ENST00000529517.1
|
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr15_-_90437195 | 0.10 |
ENST00000560940.1
ENST00000558011.1 |
AP3S2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr20_+_44035200 | 0.10 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr20_+_55926625 | 0.10 |
ENST00000452119.1
|
RAE1
|
ribonucleic acid export 1 |
| chr10_-_126849626 | 0.09 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr19_+_10217270 | 0.09 |
ENST00000446223.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr8_-_11325047 | 0.09 |
ENST00000531804.1
|
FAM167A
|
family with sequence similarity 167, member A |
| chr9_-_133814527 | 0.09 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr15_+_38544476 | 0.09 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr16_+_66400533 | 0.09 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr19_+_1000418 | 0.09 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr3_-_79816965 | 0.09 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr5_-_39425068 | 0.09 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_-_1344999 | 0.09 |
ENST00000320927.6
|
CLPTM1L
|
CLPTM1-like |
| chr2_+_25016282 | 0.09 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr8_+_144816303 | 0.09 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr4_+_75311019 | 0.09 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr22_-_29137771 | 0.09 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr11_-_123612319 | 0.09 |
ENST00000526252.1
ENST00000530393.1 ENST00000533463.1 ENST00000336139.4 ENST00000529691.1 ENST00000528306.1 |
ZNF202
|
zinc finger protein 202 |
| chr18_+_33709834 | 0.09 |
ENST00000358232.6
ENST00000351393.6 ENST00000442325.2 ENST00000423854.2 ENST00000350494.6 ENST00000542824.1 |
ELP2
|
elongator acetyltransferase complex subunit 2 |
| chr2_+_217277466 | 0.08 |
ENST00000358207.5
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr2_-_70417827 | 0.08 |
ENST00000457952.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr10_-_99161033 | 0.08 |
ENST00000315563.6
ENST00000370992.4 ENST00000414986.1 |
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr17_+_21187976 | 0.08 |
ENST00000342679.4
|
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr1_-_8938736 | 0.08 |
ENST00000234590.4
|
ENO1
|
enolase 1, (alpha) |
| chr1_-_43833628 | 0.08 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr14_+_23564700 | 0.08 |
ENST00000554203.1
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chr10_-_69834973 | 0.08 |
ENST00000395187.2
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr15_-_90233907 | 0.08 |
ENST00000561224.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr5_+_89770664 | 0.08 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr6_-_33290580 | 0.08 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr2_+_192110199 | 0.08 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chr6_-_107436473 | 0.08 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr11_-_63258601 | 0.08 |
ENST00000540857.1
ENST00000539221.1 ENST00000301790.4 |
HRASLS5
|
HRAS-like suppressor family, member 5 |
| chr3_-_130745571 | 0.08 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr15_+_74908228 | 0.08 |
ENST00000566126.1
|
CLK3
|
CDC-like kinase 3 |
| chr19_+_10216899 | 0.08 |
ENST00000428358.1
ENST00000393796.4 ENST00000253107.7 ENST00000556468.1 ENST00000393793.1 |
PPAN-P2RY11
PPAN
|
PPAN-P2RY11 readthrough peter pan homolog (Drosophila) |
| chr12_+_21590549 | 0.08 |
ENST00000545178.1
ENST00000240651.9 |
PYROXD1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr16_+_50775948 | 0.08 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr12_-_48213568 | 0.08 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr1_-_11865982 | 0.08 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chrX_-_140271249 | 0.07 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1 |
| chr10_-_16859361 | 0.07 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr12_+_58148842 | 0.07 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chrX_+_55478538 | 0.07 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr13_-_114567034 | 0.07 |
ENST00000327773.6
ENST00000357389.3 |
GAS6
|
growth arrest-specific 6 |
| chr17_+_41561317 | 0.07 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr5_-_892648 | 0.07 |
ENST00000483173.1
ENST00000435709.2 |
BRD9
|
bromodomain containing 9 |
| chr1_+_35225339 | 0.07 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr10_-_16859442 | 0.07 |
ENST00000602389.1
ENST00000345264.5 |
RSU1
|
Ras suppressor protein 1 |
| chr19_+_50084561 | 0.07 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr17_-_76274572 | 0.07 |
ENST00000374945.1
|
RP11-219G17.4
|
RP11-219G17.4 |
| chr20_+_35201993 | 0.07 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr19_-_10530784 | 0.07 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr6_+_3000057 | 0.07 |
ENST00000397717.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr16_+_28889703 | 0.07 |
ENST00000357084.3
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr3_+_71803201 | 0.07 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr10_-_69835001 | 0.07 |
ENST00000513996.1
ENST00000412272.2 ENST00000395198.3 ENST00000492996.2 |
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr16_-_66968265 | 0.07 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr4_+_75310851 | 0.07 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr1_-_43855444 | 0.07 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr19_-_6110474 | 0.07 |
ENST00000587181.1
ENST00000587321.1 ENST00000586806.1 ENST00000589742.1 ENST00000592546.1 ENST00000303657.5 |
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr2_+_192109911 | 0.07 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chr1_+_10003486 | 0.07 |
ENST00000403197.1
ENST00000377205.1 |
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr17_+_4843679 | 0.07 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr2_+_113816685 | 0.07 |
ENST00000393200.2
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr19_-_6110555 | 0.07 |
ENST00000593241.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr6_-_86352642 | 0.07 |
ENST00000355238.6
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_-_155658518 | 0.07 |
ENST00000404643.1
ENST00000359205.5 ENST00000407221.1 |
YY1AP1
|
YY1 associated protein 1 |
| chr17_-_19266045 | 0.07 |
ENST00000395616.3
|
B9D1
|
B9 protein domain 1 |
| chr5_-_1345199 | 0.07 |
ENST00000320895.5
|
CLPTM1L
|
CLPTM1-like |
| chr11_+_131781290 | 0.07 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr6_+_122793058 | 0.07 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr14_+_52327109 | 0.07 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr4_-_187647773 | 0.07 |
ENST00000509647.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr19_+_47759716 | 0.07 |
ENST00000221922.6
|
CCDC9
|
coiled-coil domain containing 9 |
| chr10_+_114710211 | 0.06 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr4_+_75480629 | 0.06 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr19_-_55658281 | 0.06 |
ENST00000585321.2
ENST00000587465.2 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr3_+_57741957 | 0.06 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr1_-_202776392 | 0.06 |
ENST00000235790.4
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr16_+_69373661 | 0.06 |
ENST00000254941.6
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr19_-_55658687 | 0.06 |
ENST00000593046.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr4_+_2470664 | 0.06 |
ENST00000314289.8
ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4
|
ring finger protein 4 |
| chr19_+_51226648 | 0.06 |
ENST00000599973.1
|
CLEC11A
|
C-type lectin domain family 11, member A |
| chr12_-_51611477 | 0.06 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr5_+_65892174 | 0.06 |
ENST00000404260.3
ENST00000403625.2 ENST00000406374.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_53693466 | 0.06 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr2_+_24346324 | 0.06 |
ENST00000407625.1
ENST00000420135.2 |
FAM228B
|
family with sequence similarity 228, member B |
| chr20_-_33735070 | 0.06 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr20_+_10199468 | 0.06 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr7_+_48128854 | 0.06 |
ENST00000436673.1
ENST00000429491.2 |
UPP1
|
uridine phosphorylase 1 |
| chr11_+_236540 | 0.06 |
ENST00000532097.1
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr22_-_50963976 | 0.06 |
ENST00000252785.3
ENST00000395693.3 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr15_-_43663214 | 0.06 |
ENST00000561661.1
|
ZSCAN29
|
zinc finger and SCAN domain containing 29 |
| chr16_+_2732476 | 0.06 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr14_+_56584414 | 0.06 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr20_+_43104508 | 0.06 |
ENST00000262605.4
ENST00000372904.3 |
TTPAL
|
tocopherol (alpha) transfer protein-like |
| chr4_-_54518619 | 0.06 |
ENST00000507168.1
ENST00000510143.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr16_+_50775971 | 0.06 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr2_+_191513789 | 0.06 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr10_+_91461413 | 0.06 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr19_-_44809121 | 0.06 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr1_+_110163709 | 0.06 |
ENST00000369840.2
ENST00000527846.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr1_-_157108130 | 0.06 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr18_-_34408693 | 0.06 |
ENST00000587382.1
ENST00000589049.1 ENST00000587129.1 |
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr16_-_11492366 | 0.06 |
ENST00000595360.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr12_+_53693812 | 0.06 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr16_+_28943260 | 0.06 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr5_-_39425290 | 0.06 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr22_-_30968839 | 0.06 |
ENST00000445645.1
ENST00000416358.1 ENST00000423371.1 ENST00000411821.1 ENST00000448604.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr18_+_3262415 | 0.06 |
ENST00000581193.1
ENST00000400175.5 |
MYL12B
|
myosin, light chain 12B, regulatory |
| chr7_-_2354099 | 0.06 |
ENST00000222990.3
|
SNX8
|
sorting nexin 8 |
| chr10_-_49813090 | 0.06 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr4_+_37978891 | 0.06 |
ENST00000446803.2
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr11_+_46722368 | 0.06 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr22_+_32340447 | 0.06 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
Network of associatons between targets according to the STRING database.
First level regulatory network of EP300
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.1 | 0.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.0 | 0.4 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0035938 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0044053 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.2 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.0 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.0 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |