Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for EN2_GBX2_LBX2
Z-value: 0.20
Transcription factors associated with EN2_GBX2_LBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
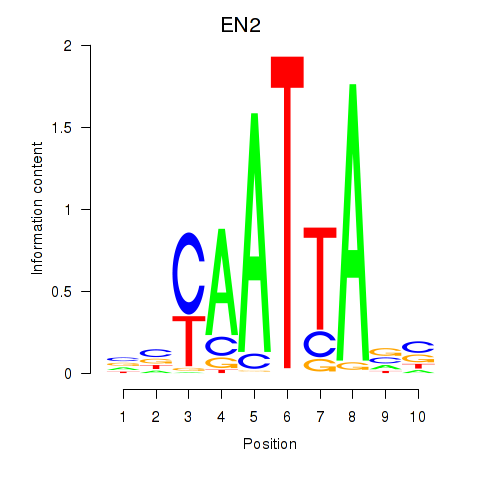
EN2
|
ENSG00000164778.4 | engrailed homeobox 2 |
|
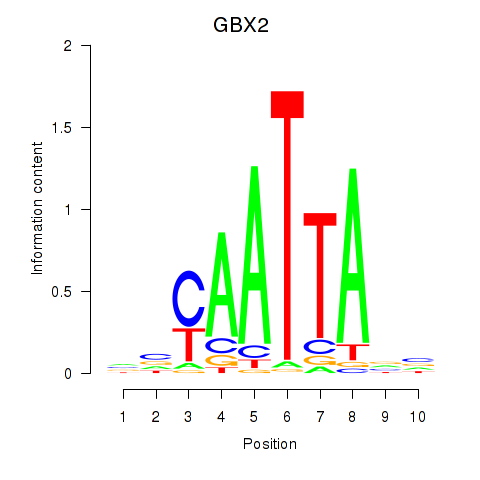
GBX2
|
ENSG00000168505.6 | gastrulation brain homeobox 2 |
|
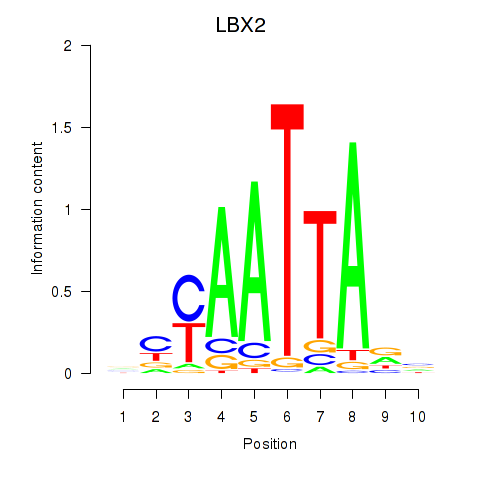
LBX2
|
ENSG00000179528.11 | ladybird homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LBX2 | hg19_v2_chr2_-_74730430_74730451 | -0.44 | 3.8e-01 | Click! |
| EN2 | hg19_v2_chr7_+_155250824_155250824 | -0.26 | 6.2e-01 | Click! |
| GBX2 | hg19_v2_chr2_-_237076992_237077012 | -0.16 | 7.6e-01 | Click! |
Activity profile of EN2_GBX2_LBX2 motif
Sorted Z-values of EN2_GBX2_LBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_77427676 | 0.27 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr16_+_53133070 | 0.14 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr6_-_31107127 | 0.13 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr4_-_36245561 | 0.13 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_+_77428066 | 0.10 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr1_+_84767289 | 0.10 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr18_+_32173276 | 0.09 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr19_-_14064114 | 0.09 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr7_+_77428149 | 0.08 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr14_-_51027838 | 0.08 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_-_234667504 | 0.08 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr18_-_33709268 | 0.08 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr5_-_95297534 | 0.08 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr12_+_28410128 | 0.08 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_62901968 | 0.07 |
ENST00000452143.1
ENST00000442679.1 ENST00000371146.1 |
USP1
|
ubiquitin specific peptidase 1 |
| chr12_-_89746173 | 0.07 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr19_-_59084647 | 0.07 |
ENST00000594234.1
ENST00000596039.1 |
MZF1
|
myeloid zinc finger 1 |
| chr5_-_115890554 | 0.06 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_-_73460334 | 0.06 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr2_-_61697862 | 0.06 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr1_-_151762943 | 0.06 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr15_-_55562451 | 0.06 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr17_-_71223839 | 0.05 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr11_-_118023594 | 0.05 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chrX_-_46187069 | 0.05 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr2_+_17997763 | 0.05 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr15_+_96904487 | 0.04 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr14_-_73360796 | 0.04 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chrX_+_95939638 | 0.04 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr4_+_169013666 | 0.04 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chrX_+_154444643 | 0.04 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr15_+_58430368 | 0.04 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr11_-_31531121 | 0.04 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr15_-_37393406 | 0.04 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr3_+_172468749 | 0.04 |
ENST00000366254.2
ENST00000415665.1 ENST00000438041.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr8_+_92261516 | 0.04 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr17_-_74163159 | 0.04 |
ENST00000591615.1
|
RNF157
|
ring finger protein 157 |
| chr3_+_130569429 | 0.03 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr3_+_172468505 | 0.03 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr5_+_125758865 | 0.03 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chrX_+_47077680 | 0.03 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr1_-_151762900 | 0.03 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr12_+_52695617 | 0.03 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr3_+_111718173 | 0.03 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chrX_+_100353153 | 0.03 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr1_-_149785236 | 0.03 |
ENST00000331491.1
|
HIST2H3D
|
histone cluster 2, H3d |
| chr11_-_33913708 | 0.03 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr2_+_149402989 | 0.03 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_-_111424506 | 0.03 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr14_+_32798547 | 0.02 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr6_-_111927449 | 0.02 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chrX_+_95939711 | 0.02 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr12_-_52828147 | 0.02 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr7_-_27169801 | 0.02 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr1_-_152131703 | 0.02 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr12_+_64798826 | 0.02 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr6_-_18264406 | 0.02 |
ENST00000515742.1
|
DEK
|
DEK oncogene |
| chr17_+_47448102 | 0.02 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr7_+_74072288 | 0.02 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr6_-_32157947 | 0.02 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr4_-_74486109 | 0.02 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_+_167453493 | 0.02 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr1_-_118472171 | 0.02 |
ENST00000369442.3
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr15_+_80351910 | 0.02 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chrX_+_129473916 | 0.02 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr3_-_178984759 | 0.02 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr18_-_53019208 | 0.02 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr3_+_133292759 | 0.02 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr14_-_82000140 | 0.02 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr6_-_111927062 | 0.02 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr3_+_119814070 | 0.02 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr11_+_75526212 | 0.02 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr1_-_68698197 | 0.02 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr22_+_41777927 | 0.02 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr12_-_57328187 | 0.02 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chrX_+_134654540 | 0.02 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr4_+_183065793 | 0.02 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr15_-_55563072 | 0.02 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_+_172468472 | 0.02 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr1_-_2461684 | 0.02 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr12_+_53662073 | 0.02 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr14_+_32798462 | 0.02 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr16_+_53164833 | 0.02 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr15_-_64673665 | 0.02 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr7_-_92777606 | 0.02 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr3_+_111717511 | 0.02 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr5_+_125758813 | 0.02 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr16_+_53164956 | 0.01 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_80912059 | 0.01 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr11_+_57435441 | 0.01 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr5_+_31193847 | 0.01 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr9_+_2159850 | 0.01 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_-_31088214 | 0.01 |
ENST00000376288.2
|
CDSN
|
corneodesmosin |
| chr1_-_232651312 | 0.01 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr4_-_89205879 | 0.01 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr12_-_56694142 | 0.01 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr9_+_12693336 | 0.01 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr1_+_153190053 | 0.01 |
ENST00000368744.3
|
PRR9
|
proline rich 9 |
| chr20_-_18447667 | 0.01 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr4_-_74486347 | 0.01 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_+_135502501 | 0.01 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr15_+_89631381 | 0.01 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr2_+_171034646 | 0.01 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr2_-_157189180 | 0.01 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_+_210501589 | 0.01 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr12_+_7014126 | 0.01 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr1_-_23670817 | 0.01 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chrX_+_123097014 | 0.01 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr19_-_10121144 | 0.01 |
ENST00000264828.3
|
COL5A3
|
collagen, type V, alpha 3 |
| chr1_+_62439037 | 0.01 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_-_23670813 | 0.01 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chrX_-_53461288 | 0.01 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr1_-_23670752 | 0.01 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr3_+_152879985 | 0.01 |
ENST00000323534.2
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr8_-_128231299 | 0.01 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr3_+_111718036 | 0.01 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr17_-_47045949 | 0.01 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr2_+_242089833 | 0.01 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr14_+_104182105 | 0.01 |
ENST00000311141.2
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr14_-_78083112 | 0.01 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr2_+_47630108 | 0.01 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr18_+_32402321 | 0.01 |
ENST00000587723.1
|
DTNA
|
dystrobrevin, alpha |
| chr15_-_31393910 | 0.01 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr7_-_83278322 | 0.01 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr18_-_31803169 | 0.01 |
ENST00000590712.1
|
NOL4
|
nucleolar protein 4 |
| chr19_+_48949087 | 0.01 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr4_+_71587669 | 0.01 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr2_+_86668464 | 0.01 |
ENST00000409064.1
|
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr2_+_105654441 | 0.01 |
ENST00000258455.3
|
MRPS9
|
mitochondrial ribosomal protein S9 |
| chr8_+_9413410 | 0.01 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr4_-_74486217 | 0.01 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr2_+_47630255 | 0.01 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr6_-_139613269 | 0.01 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr12_-_16760021 | 0.01 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_68698222 | 0.01 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr3_+_111717600 | 0.01 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr4_-_69111401 | 0.01 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr4_-_89205705 | 0.01 |
ENST00000295908.7
ENST00000510548.2 ENST00000508256.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr3_+_115342349 | 0.01 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr14_-_74551172 | 0.01 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr17_+_7792101 | 0.01 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr20_-_50722183 | 0.01 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr19_-_40931891 | 0.00 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr6_+_39760129 | 0.00 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr1_-_157746909 | 0.00 |
ENST00000392274.3
ENST00000361516.3 ENST00000368181.4 |
FCRL2
|
Fc receptor-like 2 |
| chr19_+_34287751 | 0.00 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr16_+_53412368 | 0.00 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr3_-_112360116 | 0.00 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr12_+_53662110 | 0.00 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr3_+_156799587 | 0.00 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr10_-_99447024 | 0.00 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr4_-_57547870 | 0.00 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr15_+_58430567 | 0.00 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr5_+_145826867 | 0.00 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr1_+_152943122 | 0.00 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr4_-_174255536 | 0.00 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr7_-_107642348 | 0.00 |
ENST00000393561.1
|
LAMB1
|
laminin, beta 1 |
| chr18_+_55888767 | 0.00 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr9_-_133769225 | 0.00 |
ENST00000343079.1
|
QRFP
|
pyroglutamylated RFamide peptide |
| chr21_-_43816052 | 0.00 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr15_-_64673630 | 0.00 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr2_+_158114051 | 0.00 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chrX_-_53461305 | 0.00 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr4_-_143226979 | 0.00 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr4_-_143227088 | 0.00 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr17_-_39203519 | 0.00 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr4_-_174255400 | 0.00 |
ENST00000506267.1
|
HMGB2
|
high mobility group box 2 |
| chr9_+_116225999 | 0.00 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr2_+_65216462 | 0.00 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_+_141303373 | 0.00 |
ENST00000432126.2
ENST00000194118.4 |
KIAA0141
|
KIAA0141 |
| chr17_-_48785216 | 0.00 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr1_+_110881945 | 0.00 |
ENST00000602849.1
ENST00000487146.2 |
RBM15
|
RNA binding motif protein 15 |
| chr17_-_10017864 | 0.00 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr2_-_224467093 | 0.00 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr6_-_116833500 | 0.00 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr7_-_137028534 | 0.00 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr10_+_99205959 | 0.00 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr4_-_174256276 | 0.00 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr3_-_157217328 | 0.00 |
ENST00000392832.2
ENST00000543418.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr14_+_104182061 | 0.00 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr6_-_72130472 | 0.00 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr4_-_39979576 | 0.00 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr3_-_27764190 | 0.00 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr10_-_56561022 | 0.00 |
ENST00000373965.2
ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15
|
protocadherin-related 15 |
| chr19_+_51728316 | 0.00 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr11_-_71823715 | 0.00 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EN2_GBX2_LBX2
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |