Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
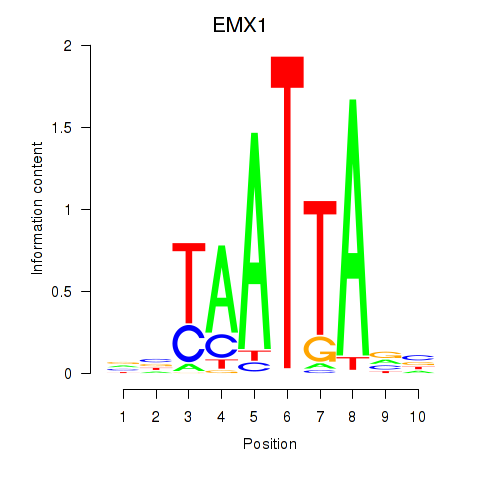
Results for EMX1
Z-value: 0.78
Transcription factors associated with EMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX1
|
ENSG00000135638.9 | empty spiracles homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX1 | hg19_v2_chr2_+_73144604_73144654 | 0.20 | 7.0e-01 | Click! |
Activity profile of EMX1 motif
Sorted Z-values of EMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_36245561 | 0.52 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_-_75488984 | 0.52 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chrX_+_13671225 | 0.51 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr6_-_31107127 | 0.43 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr9_-_3469181 | 0.41 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr11_-_111649074 | 0.40 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr7_-_77427676 | 0.40 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr8_+_105235572 | 0.38 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_+_94451574 | 0.38 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr8_+_97773202 | 0.35 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr1_+_62439037 | 0.34 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr5_-_24645078 | 0.33 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr5_+_135394840 | 0.32 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr20_-_271304 | 0.32 |
ENST00000400269.3
ENST00000360321.2 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr6_+_148593425 | 0.32 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr4_+_175204818 | 0.32 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr2_-_136678123 | 0.31 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr13_-_36050819 | 0.29 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr3_+_121774202 | 0.29 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr12_-_118796910 | 0.28 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr7_+_77428066 | 0.28 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr4_-_69111401 | 0.28 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr7_+_77428149 | 0.28 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr5_-_142077569 | 0.27 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr2_-_203735586 | 0.26 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr6_+_139135648 | 0.26 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr4_-_74486109 | 0.26 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_+_44229846 | 0.26 |
ENST00000551577.1
ENST00000266534.3 |
TMEM117
|
transmembrane protein 117 |
| chr17_-_46799872 | 0.26 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr3_+_174158732 | 0.25 |
ENST00000434257.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr1_-_68698197 | 0.25 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr5_+_115177178 | 0.25 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr5_-_137374288 | 0.25 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr10_-_96829246 | 0.25 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr1_+_202317815 | 0.25 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr4_+_146402346 | 0.25 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr14_-_73360796 | 0.25 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr6_-_10435032 | 0.25 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr7_+_23719749 | 0.24 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr4_-_89442940 | 0.24 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr2_-_227050079 | 0.24 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr12_+_107712173 | 0.24 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr4_-_147443043 | 0.24 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr10_-_93392811 | 0.24 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr4_+_169013666 | 0.23 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr7_+_12726623 | 0.23 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr20_+_12989596 | 0.23 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr6_-_109702885 | 0.23 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr7_+_6655225 | 0.22 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chrX_+_102024075 | 0.22 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr2_-_102003987 | 0.22 |
ENST00000324768.5
|
CREG2
|
cellular repressor of E1A-stimulated genes 2 |
| chr17_+_48823975 | 0.22 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr8_-_17533838 | 0.22 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr7_-_27169801 | 0.20 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr4_+_183065793 | 0.20 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr2_+_171034646 | 0.20 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr5_+_36608422 | 0.20 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_-_15815626 | 0.20 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr4_-_89205879 | 0.20 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr7_+_130126165 | 0.20 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr11_-_104905840 | 0.20 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr1_-_68698222 | 0.20 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr7_+_116660246 | 0.19 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr14_+_67831576 | 0.19 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr3_-_180397256 | 0.19 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr12_-_118796971 | 0.19 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr10_-_23633720 | 0.19 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr14_-_70883708 | 0.19 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr19_+_37862054 | 0.19 |
ENST00000483919.1
ENST00000588911.1 ENST00000436120.2 ENST00000587349.1 |
ZNF527
|
zinc finger protein 527 |
| chr8_+_70404996 | 0.19 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr7_-_83278322 | 0.19 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr17_-_39123144 | 0.19 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr3_+_156799587 | 0.19 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr7_-_111032971 | 0.19 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr6_+_130339710 | 0.19 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr19_-_14064114 | 0.18 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr2_-_207082748 | 0.18 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr10_-_128110441 | 0.18 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr4_+_105828492 | 0.18 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr14_-_51027838 | 0.18 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr19_-_12889226 | 0.18 |
ENST00000589400.1
ENST00000590839.1 ENST00000592079.1 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr3_-_123339343 | 0.18 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr14_+_32798547 | 0.18 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr8_+_42396274 | 0.18 |
ENST00000438528.3
|
SMIM19
|
small integral membrane protein 19 |
| chr20_+_12989895 | 0.18 |
ENST00000450297.1
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr11_-_96076334 | 0.18 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr12_+_16500599 | 0.18 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr20_+_12989822 | 0.17 |
ENST00000378194.4
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr12_+_16500571 | 0.17 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr10_+_91461337 | 0.17 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr3_-_151034734 | 0.17 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_+_53751445 | 0.17 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr1_-_151965048 | 0.16 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr15_+_96904487 | 0.16 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr7_+_93652116 | 0.16 |
ENST00000415536.1
|
AC003092.1
|
AC003092.1 |
| chr6_+_153552455 | 0.16 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr19_-_58951496 | 0.16 |
ENST00000254166.3
|
ZNF132
|
zinc finger protein 132 |
| chr12_+_16500037 | 0.16 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chrX_-_46187069 | 0.16 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr16_-_53737722 | 0.16 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr12_-_50616382 | 0.16 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr5_-_148929848 | 0.16 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chrX_+_134654540 | 0.16 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr8_-_90996837 | 0.16 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr16_-_66764119 | 0.16 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr7_+_36450169 | 0.16 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr12_+_64798826 | 0.16 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr15_-_55562451 | 0.15 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_+_86525299 | 0.15 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr16_-_66864806 | 0.15 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr14_-_21737551 | 0.15 |
ENST00000554891.1
ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr20_-_23860373 | 0.15 |
ENST00000304710.4
|
CST5
|
cystatin D |
| chr5_-_74162605 | 0.15 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr5_+_36606700 | 0.15 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr5_+_36608280 | 0.15 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_-_38670897 | 0.15 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr1_-_151798546 | 0.15 |
ENST00000356728.6
|
RORC
|
RAR-related orphan receptor C |
| chr12_+_41136144 | 0.15 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr10_+_5454505 | 0.15 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr1_-_151762943 | 0.14 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr10_+_111765562 | 0.14 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr3_+_152552685 | 0.14 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr16_-_53737795 | 0.14 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr17_-_74163159 | 0.14 |
ENST00000591615.1
|
RNF157
|
ring finger protein 157 |
| chr11_-_107729887 | 0.14 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_+_169633310 | 0.14 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_-_42878101 | 0.14 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr12_-_25348007 | 0.14 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr6_-_111804905 | 0.14 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr14_+_85994943 | 0.14 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr7_-_111424462 | 0.14 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr11_+_112041253 | 0.14 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr1_+_236958554 | 0.14 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chrX_-_13835147 | 0.13 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr7_+_130126012 | 0.13 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr1_-_2461684 | 0.13 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr4_-_74486347 | 0.13 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chrX_+_22050546 | 0.13 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr1_+_202317855 | 0.13 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_-_28571015 | 0.13 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr9_+_26956371 | 0.13 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr7_+_115862858 | 0.13 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr5_-_95297534 | 0.13 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chrX_+_105855160 | 0.13 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr2_+_197577841 | 0.13 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chrX_-_106243451 | 0.13 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr11_+_100862811 | 0.13 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr11_-_118134997 | 0.13 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr11_+_27015628 | 0.13 |
ENST00000318627.2
|
FIBIN
|
fin bud initiation factor homolog (zebrafish) |
| chr11_-_118023490 | 0.13 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr1_-_157746909 | 0.13 |
ENST00000392274.3
ENST00000361516.3 ENST00000368181.4 |
FCRL2
|
Fc receptor-like 2 |
| chr1_-_205719295 | 0.12 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr18_+_32173276 | 0.12 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr2_+_29320571 | 0.12 |
ENST00000401605.1
ENST00000401617.2 |
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr8_-_116504448 | 0.12 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr14_-_21737610 | 0.12 |
ENST00000320084.7
ENST00000449098.1 ENST00000336053.6 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr11_+_33061543 | 0.12 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr12_+_106751436 | 0.12 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr13_-_41593425 | 0.12 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr5_+_68860949 | 0.12 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chrX_-_107334790 | 0.12 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr15_-_72565340 | 0.12 |
ENST00000568360.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr3_+_186353756 | 0.12 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr15_-_65903407 | 0.12 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr3_-_168865522 | 0.12 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr5_-_159846066 | 0.12 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr7_+_12727250 | 0.12 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr9_+_70856899 | 0.12 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr12_+_128399965 | 0.11 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr4_+_129732419 | 0.11 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr14_+_52313833 | 0.11 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr10_-_27529486 | 0.11 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr17_+_33914424 | 0.11 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr11_+_77532155 | 0.11 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr12_+_32638897 | 0.11 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr12_-_22063787 | 0.11 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr19_-_56110859 | 0.11 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr1_-_109935819 | 0.11 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr2_+_179184955 | 0.11 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr1_+_160370344 | 0.11 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr10_-_116418053 | 0.11 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr8_-_101963482 | 0.11 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr16_+_57279004 | 0.11 |
ENST00000219204.3
|
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr9_+_109685630 | 0.11 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr5_-_16509101 | 0.11 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr17_+_79495397 | 0.11 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr18_-_33709268 | 0.11 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr4_-_74486217 | 0.11 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_-_88974236 | 0.11 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr2_-_99871570 | 0.11 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr15_+_86087267 | 0.11 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chrX_-_118986911 | 0.10 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr3_+_107364769 | 0.10 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_+_210501589 | 0.10 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr9_+_26956474 | 0.10 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr10_-_71169031 | 0.10 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr4_+_146539415 | 0.10 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr17_-_55911970 | 0.10 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr14_-_23652849 | 0.10 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr13_+_36050881 | 0.10 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.4 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 | 0.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.2 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.3 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.3 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) regulation of penile erection(GO:0060405) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0035106 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.0 | 0.6 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.3 | GO:2000544 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:1903717 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.0 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.3 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.0 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |