Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
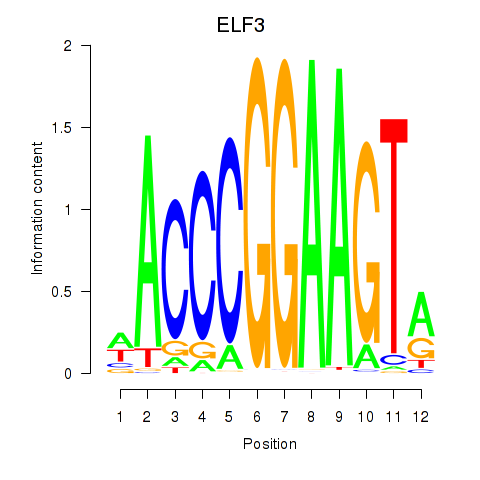
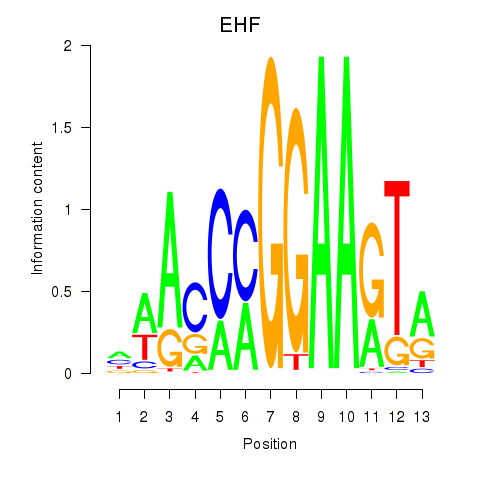
Results for ELF3_EHF
Z-value: 0.82
Transcription factors associated with ELF3_EHF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ELF3
|
ENSG00000163435.11 | E74 like ETS transcription factor 3 |
|
EHF
|
ENSG00000135373.8 | ETS homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EHF | hg19_v2_chr11_+_34642656_34642682 | -0.54 | 2.7e-01 | Click! |
| ELF3 | hg19_v2_chr1_+_201979645_201979721 | 0.49 | 3.2e-01 | Click! |
Activity profile of ELF3_EHF motif
Sorted Z-values of ELF3_EHF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_242913327 | 0.93 |
ENST00000426962.1
|
AC093642.3
|
AC093642.3 |
| chr8_+_182422 | 0.66 |
ENST00000518414.1
ENST00000521270.1 ENST00000518320.2 |
ZNF596
|
zinc finger protein 596 |
| chr19_+_41497178 | 0.47 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr13_-_114103443 | 0.42 |
ENST00000356501.4
ENST00000413169.2 |
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr5_-_110074603 | 0.38 |
ENST00000515278.2
|
TMEM232
|
transmembrane protein 232 |
| chr12_-_58329888 | 0.38 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr5_+_172484377 | 0.35 |
ENST00000523161.1
|
CREBRF
|
CREB3 regulatory factor |
| chr1_-_12679171 | 0.35 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr19_-_45004556 | 0.33 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr10_-_22292675 | 0.31 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr10_-_98031310 | 0.30 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr8_+_182368 | 0.30 |
ENST00000522866.1
ENST00000398612.1 |
ZNF596
|
zinc finger protein 596 |
| chr19_+_56813305 | 0.30 |
ENST00000593151.1
|
AC006116.20
|
Uncharacterized protein |
| chr2_-_241500168 | 0.29 |
ENST00000443318.1
ENST00000411765.1 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr12_-_123215306 | 0.29 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr14_-_69864993 | 0.27 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chr19_+_52956798 | 0.27 |
ENST00000421239.2
|
ZNF578
|
zinc finger protein 578 |
| chr11_+_111789580 | 0.24 |
ENST00000278601.5
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr22_+_27068766 | 0.23 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr4_-_140005341 | 0.22 |
ENST00000379549.2
ENST00000512627.1 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr16_+_88636875 | 0.22 |
ENST00000569435.1
|
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr2_+_66662249 | 0.22 |
ENST00000560281.2
|
MEIS1
|
Meis homeobox 1 |
| chr16_-_25122735 | 0.22 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr3_-_47324079 | 0.21 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr3_-_52322019 | 0.21 |
ENST00000463624.1
|
WDR82
|
WD repeat domain 82 |
| chrX_-_80457385 | 0.21 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr6_-_11779174 | 0.21 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_-_146697185 | 0.21 |
ENST00000533174.1
ENST00000254090.4 |
FMO5
|
flavin containing monooxygenase 5 |
| chr3_-_42003613 | 0.20 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr3_-_20227720 | 0.20 |
ENST00000412997.1
|
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr6_-_11779403 | 0.20 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr18_+_43246028 | 0.20 |
ENST00000589658.1
|
SLC14A2
|
solute carrier family 14 (urea transporter), member 2 |
| chr9_-_86571628 | 0.20 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr22_+_27068704 | 0.20 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr3_-_180397256 | 0.20 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr16_-_25122785 | 0.20 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr19_-_55791431 | 0.20 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr7_+_106415457 | 0.19 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr19_-_12708113 | 0.19 |
ENST00000440366.1
|
ZNF490
|
zinc finger protein 490 |
| chr3_+_119217422 | 0.19 |
ENST00000466984.1
|
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr1_-_151882031 | 0.19 |
ENST00000489410.1
|
THEM4
|
thioesterase superfamily member 4 |
| chr16_-_2205352 | 0.19 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr1_+_43124087 | 0.19 |
ENST00000304979.3
ENST00000372550.1 ENST00000440068.1 |
PPIH
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr4_-_153700864 | 0.19 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr12_-_12509929 | 0.19 |
ENST00000381800.2
|
LOH12CR2
|
loss of heterozygosity, 12, chromosomal region 2 (non-protein coding) |
| chr4_+_37828255 | 0.18 |
ENST00000381967.4
ENST00000544359.1 ENST00000537241.1 |
PGM2
|
phosphoglucomutase 2 |
| chr3_-_171527560 | 0.18 |
ENST00000331659.2
|
PP13439
|
PP13439 |
| chr1_+_171283331 | 0.18 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr7_+_33168856 | 0.18 |
ENST00000432983.1
|
BBS9
|
Bardet-Biedl syndrome 9 |
| chr15_-_90233907 | 0.17 |
ENST00000561224.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr1_-_86861660 | 0.17 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr6_+_111303218 | 0.17 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr9_+_100174344 | 0.17 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr8_-_131028782 | 0.17 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr9_+_71944241 | 0.17 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr2_-_99485825 | 0.16 |
ENST00000423771.1
|
KIAA1211L
|
KIAA1211-like |
| chr2_-_175351744 | 0.16 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr15_-_90233866 | 0.16 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chrX_+_129473859 | 0.16 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr15_-_64648273 | 0.16 |
ENST00000607537.1
ENST00000303052.7 ENST00000303032.6 |
CSNK1G1
|
casein kinase 1, gamma 1 |
| chr19_-_55791563 | 0.16 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr21_-_33984865 | 0.16 |
ENST00000458138.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr12_-_118796910 | 0.16 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr17_+_34958001 | 0.16 |
ENST00000250156.7
|
MRM1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr16_+_4845379 | 0.16 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr12_-_6602955 | 0.16 |
ENST00000543703.1
|
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr3_-_139108463 | 0.16 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr6_+_49431073 | 0.16 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr2_+_136499287 | 0.15 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr3_+_42977846 | 0.15 |
ENST00000383748.4
|
KRBOX1
|
KRAB box domain containing 1 |
| chr5_+_137203541 | 0.15 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr7_-_150020216 | 0.15 |
ENST00000477367.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr17_-_76837499 | 0.15 |
ENST00000592275.1
|
USP36
|
ubiquitin specific peptidase 36 |
| chr1_-_197744763 | 0.15 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr14_-_71107921 | 0.14 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr1_+_53662101 | 0.14 |
ENST00000371486.3
|
CPT2
|
carnitine palmitoyltransferase 2 |
| chr1_+_95286151 | 0.14 |
ENST00000467909.1
ENST00000422520.2 ENST00000532427.1 |
SLC44A3
|
solute carrier family 44, member 3 |
| chr12_+_7072354 | 0.14 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr1_+_213224572 | 0.14 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr3_+_15469058 | 0.14 |
ENST00000432764.2
|
EAF1
|
ELL associated factor 1 |
| chrX_+_47092791 | 0.14 |
ENST00000377080.3
|
USP11
|
ubiquitin specific peptidase 11 |
| chr1_+_198126209 | 0.14 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr14_-_31926595 | 0.14 |
ENST00000547378.1
|
RP11-176H8.1
|
Uncharacterized protein |
| chr12_+_21679220 | 0.14 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr2_+_217363559 | 0.14 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr7_-_150020814 | 0.14 |
ENST00000477871.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr12_+_6602517 | 0.14 |
ENST00000315579.5
ENST00000539714.1 |
NCAPD2
|
non-SMC condensin I complex, subunit D2 |
| chr11_+_32605350 | 0.14 |
ENST00000531120.1
ENST00000524896.1 ENST00000323213.5 |
EIF3M
|
eukaryotic translation initiation factor 3, subunit M |
| chr3_-_52719888 | 0.14 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr1_+_46049706 | 0.13 |
ENST00000527470.1
ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr19_-_53289995 | 0.13 |
ENST00000338230.3
|
ZNF600
|
zinc finger protein 600 |
| chr16_+_19729586 | 0.13 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr19_-_55791540 | 0.13 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr2_-_73964447 | 0.13 |
ENST00000272424.5
ENST00000409716.2 ENST00000318190.7 |
TPRKB
|
TP53RK binding protein |
| chr1_+_40942887 | 0.13 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chrX_+_70586140 | 0.13 |
ENST00000276072.3
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr19_+_56146352 | 0.13 |
ENST00000592881.1
|
ZNF580
|
zinc finger protein 580 |
| chr15_+_89787180 | 0.13 |
ENST00000300027.8
ENST00000310775.7 ENST00000567891.1 ENST00000564920.1 ENST00000565255.1 ENST00000567996.1 ENST00000451393.2 ENST00000563250.1 |
FANCI
|
Fanconi anemia, complementation group I |
| chr11_+_118889142 | 0.13 |
ENST00000533632.1
|
TRAPPC4
|
trafficking protein particle complex 4 |
| chr20_+_44486246 | 0.13 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr4_-_2366525 | 0.13 |
ENST00000515169.1
ENST00000515312.1 |
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr2_+_48667983 | 0.13 |
ENST00000449090.2
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr7_+_138915102 | 0.13 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr1_+_92683467 | 0.13 |
ENST00000370375.3
ENST00000370373.2 |
C1orf146
|
chromosome 1 open reading frame 146 |
| chr14_+_65878650 | 0.12 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr8_+_100025476 | 0.12 |
ENST00000355155.1
ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast) |
| chr7_+_30589829 | 0.12 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr18_-_28742813 | 0.12 |
ENST00000257197.3
ENST00000257198.5 |
DSC1
|
desmocollin 1 |
| chr14_+_24701819 | 0.12 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr4_+_166128735 | 0.12 |
ENST00000226725.6
|
KLHL2
|
kelch-like family member 2 |
| chr10_-_98031265 | 0.12 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr2_+_48667898 | 0.12 |
ENST00000281394.4
ENST00000294952.8 |
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr2_+_203776937 | 0.12 |
ENST00000402905.3
ENST00000414490.1 ENST00000431787.1 ENST00000444724.1 ENST00000414857.1 ENST00000430899.1 ENST00000445120.1 ENST00000441569.1 ENST00000432024.1 ENST00000443740.1 ENST00000414439.1 ENST00000428585.1 ENST00000545253.1 ENST00000545262.1 ENST00000447539.1 ENST00000456821.2 ENST00000434998.1 ENST00000320443.8 |
CARF
|
calcium responsive transcription factor |
| chr8_-_131028869 | 0.12 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr15_+_23810903 | 0.12 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr10_-_14996017 | 0.12 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr5_+_180650271 | 0.12 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr3_+_119217376 | 0.12 |
ENST00000494664.1
ENST00000493694.1 |
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr4_+_165675197 | 0.12 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr21_+_17909594 | 0.12 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_-_71088797 | 0.12 |
ENST00000580557.1
ENST00000579732.1 ENST00000578620.1 ENST00000542342.2 ENST00000255559.3 ENST00000579018.1 |
SLC39A11
|
solute carrier family 39, member 11 |
| chr2_-_208890218 | 0.12 |
ENST00000457206.1
ENST00000427836.2 ENST00000389247.4 |
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3 |
| chr2_-_120124258 | 0.11 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr6_+_111580508 | 0.11 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr17_-_56065540 | 0.11 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chrX_-_107018969 | 0.11 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr2_+_66662510 | 0.11 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr5_-_140700322 | 0.11 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr9_-_77703115 | 0.11 |
ENST00000361092.4
ENST00000376808.4 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr1_-_52870059 | 0.11 |
ENST00000371566.1
|
ORC1
|
origin recognition complex, subunit 1 |
| chr4_+_86699834 | 0.11 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_-_19648416 | 0.11 |
ENST00000426645.2
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_+_100598742 | 0.11 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr16_+_84682108 | 0.11 |
ENST00000564996.1
ENST00000258157.5 ENST00000567410.1 |
KLHL36
|
kelch-like family member 36 |
| chr2_-_69870747 | 0.11 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr7_-_38948774 | 0.11 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr14_+_45366518 | 0.11 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr18_-_73967160 | 0.11 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr6_+_139349903 | 0.11 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr4_+_166128836 | 0.11 |
ENST00000511305.1
|
KLHL2
|
kelch-like family member 2 |
| chr19_-_49552363 | 0.11 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
| chr6_-_28303901 | 0.11 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr1_+_231114795 | 0.10 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr6_-_32557610 | 0.10 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr19_-_49540073 | 0.10 |
ENST00000301407.7
ENST00000601167.1 ENST00000604577.1 ENST00000591656.1 |
CGB1
CTB-60B18.6
|
chorionic gonadotropin, beta polypeptide 1 Choriogonadotropin subunit beta variant 1; Uncharacterized protein |
| chr6_+_33257427 | 0.10 |
ENST00000463584.1
|
PFDN6
|
prefoldin subunit 6 |
| chr19_+_53700340 | 0.10 |
ENST00000597550.1
ENST00000601072.1 |
CTD-2245F17.3
|
CTD-2245F17.3 |
| chr1_-_28969517 | 0.10 |
ENST00000263974.4
ENST00000373824.4 |
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr2_+_202125219 | 0.10 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr3_+_100120441 | 0.10 |
ENST00000489752.1
|
LNP1
|
leukemia NUP98 fusion partner 1 |
| chr15_-_102192567 | 0.10 |
ENST00000333202.3
ENST00000428002.2 ENST00000559107.1 ENST00000347970.3 |
TM2D3
|
TM2 domain containing 3 |
| chr11_-_62389621 | 0.10 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr14_+_75179840 | 0.10 |
ENST00000554590.1
ENST00000341162.4 ENST00000534938.2 ENST00000553615.1 |
FCF1
|
FCF1 rRNA-processing protein |
| chr19_+_45542295 | 0.10 |
ENST00000221455.3
ENST00000391953.4 ENST00000588936.1 |
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr9_-_34329198 | 0.10 |
ENST00000379166.2
ENST00000345050.2 |
KIF24
|
kinesin family member 24 |
| chr9_-_140095186 | 0.10 |
ENST00000409012.4
|
TPRN
|
taperin |
| chr1_+_204485503 | 0.10 |
ENST00000367182.3
ENST00000507825.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr20_+_29611833 | 0.10 |
ENST00000278882.3
ENST00000439954.2 ENST00000358464.4 |
FRG1B
|
FSHD region gene 1 family, member B |
| chr3_-_184429735 | 0.10 |
ENST00000317897.3
|
MAGEF1
|
melanoma antigen family F, 1 |
| chrX_-_100307076 | 0.10 |
ENST00000338687.7
ENST00000545398.1 ENST00000372931.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chr5_-_39270725 | 0.10 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr8_-_124665190 | 0.10 |
ENST00000325995.7
|
KLHL38
|
kelch-like family member 38 |
| chr15_+_80987617 | 0.10 |
ENST00000258884.4
ENST00000558464.1 |
ABHD17C
|
abhydrolase domain containing 17C |
| chr19_-_17186229 | 0.10 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chrX_-_107334750 | 0.10 |
ENST00000340200.5
ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr13_-_99852916 | 0.10 |
ENST00000426037.2
ENST00000445737.2 |
UBAC2-AS1
|
UBAC2 antisense RNA 1 |
| chr19_+_17326521 | 0.10 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr10_-_1034237 | 0.10 |
ENST00000381466.1
|
AL359878.1
|
Uncharacterized protein |
| chr2_-_241500447 | 0.10 |
ENST00000536462.1
ENST00000405002.1 ENST00000441168.1 ENST00000403283.1 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr2_-_201374781 | 0.10 |
ENST00000359878.3
ENST00000409157.1 |
KCTD18
|
potassium channel tetramerization domain containing 18 |
| chr3_-_123411191 | 0.09 |
ENST00000354792.5
ENST00000508240.1 |
MYLK
|
myosin light chain kinase |
| chr22_+_46731676 | 0.09 |
ENST00000424260.2
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr6_-_85473156 | 0.09 |
ENST00000606784.1
ENST00000606325.1 |
TBX18
|
T-box 18 |
| chr11_+_110300607 | 0.09 |
ENST00000260270.2
|
FDX1
|
ferredoxin 1 |
| chr10_+_15139394 | 0.09 |
ENST00000441850.1
|
RPP38
|
ribonuclease P/MRP 38kDa subunit |
| chr3_-_37217756 | 0.09 |
ENST00000440230.1
ENST00000421276.2 ENST00000421307.1 ENST00000354379.4 |
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr13_+_51913819 | 0.09 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr6_-_28304152 | 0.09 |
ENST00000435857.1
|
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr5_+_2752258 | 0.09 |
ENST00000334000.3
|
C5orf38
|
chromosome 5 open reading frame 38 |
| chr3_-_69370095 | 0.09 |
ENST00000473029.1
|
FRMD4B
|
FERM domain containing 4B |
| chr1_-_169337176 | 0.09 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr14_-_75593708 | 0.09 |
ENST00000557673.1
ENST00000238616.5 |
NEK9
|
NIMA-related kinase 9 |
| chr5_+_96079240 | 0.09 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr3_-_57113281 | 0.09 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr11_+_74303612 | 0.09 |
ENST00000527458.1
ENST00000532497.1 ENST00000530511.1 |
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chrX_+_70586082 | 0.09 |
ENST00000373790.4
ENST00000449580.1 ENST00000423759.1 |
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr1_+_81106951 | 0.09 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr11_-_118889323 | 0.09 |
ENST00000527673.1
|
RPS25
|
ribosomal protein S25 |
| chr10_+_101491968 | 0.09 |
ENST00000370476.5
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr19_+_55417499 | 0.09 |
ENST00000291890.4
ENST00000447255.1 ENST00000598576.1 ENST00000594765.1 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr2_-_224467093 | 0.09 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr12_-_118796971 | 0.09 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chrX_-_7895755 | 0.09 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr7_-_99149715 | 0.09 |
ENST00000449309.1
|
FAM200A
|
family with sequence similarity 200, member A |
| chr3_-_133380731 | 0.09 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr15_+_55611401 | 0.09 |
ENST00000566999.1
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr2_+_217363793 | 0.09 |
ENST00000456586.1
ENST00000598925.1 ENST00000427280.2 |
RPL37A
|
ribosomal protein L37a |
| chr4_-_168155577 | 0.09 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_+_42264322 | 0.09 |
ENST00000446571.3
ENST00000357984.3 ENST00000538716.2 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr11_-_119993734 | 0.09 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ELF3_EHF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.3 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.6 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.0 | 0.0 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0039007 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.1 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.1 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 0.1 | GO:0061570 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.0 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.0 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.0 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.0 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.0 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.0 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.3 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.2 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0016731 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.1 | GO:0032396 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.0 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.0 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.0 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.0 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.0 | GO:0004794 | L-serine ammonia-lyase activity(GO:0003941) L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |