Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
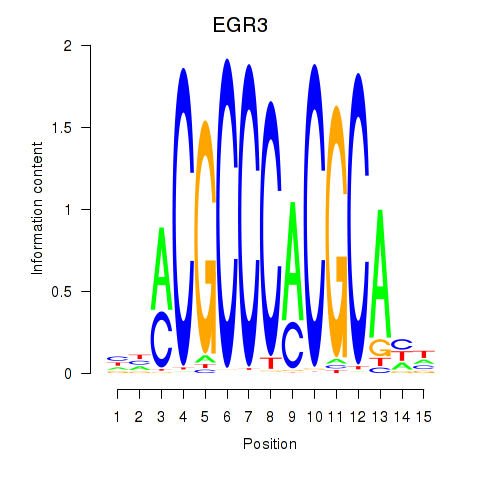
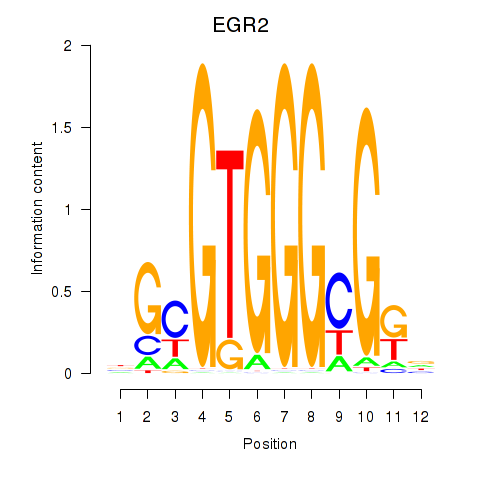
Results for EGR3_EGR2
Z-value: 0.38
Transcription factors associated with EGR3_EGR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR3
|
ENSG00000179388.8 | early growth response 3 |
|
EGR2
|
ENSG00000122877.9 | early growth response 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR2 | hg19_v2_chr10_-_64576105_64576133 | -0.96 | 2.1e-03 | Click! |
| EGR3 | hg19_v2_chr8_-_22549856_22549927 | 0.38 | 4.6e-01 | Click! |
Activity profile of EGR3_EGR2 motif
Sorted Z-values of EGR3_EGR2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_49622348 | 0.52 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr15_-_80263506 | 0.38 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr5_-_176924562 | 0.38 |
ENST00000359895.2
ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr19_+_49617581 | 0.33 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr6_+_31543334 | 0.28 |
ENST00000449264.2
|
TNF
|
tumor necrosis factor |
| chr6_+_43739697 | 0.24 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr14_-_69445793 | 0.24 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr14_-_69445968 | 0.23 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr6_+_138188551 | 0.23 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr1_+_43148059 | 0.22 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr6_+_138188351 | 0.22 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_-_14201776 | 0.19 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr6_+_138188378 | 0.19 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr2_-_27718052 | 0.18 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr8_+_32406137 | 0.18 |
ENST00000521670.1
|
NRG1
|
neuregulin 1 |
| chr1_+_43148625 | 0.18 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr22_-_30722912 | 0.18 |
ENST00000215790.7
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr1_-_16302608 | 0.18 |
ENST00000375743.4
ENST00000375733.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr3_+_101546827 | 0.17 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr15_-_75743991 | 0.17 |
ENST00000567289.1
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr22_+_37956453 | 0.17 |
ENST00000249014.4
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chrX_-_19905577 | 0.16 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr16_-_30134441 | 0.16 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr19_+_50180409 | 0.16 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr16_+_30194916 | 0.16 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chrX_-_48814810 | 0.15 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr16_-_30134524 | 0.15 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr1_-_16302565 | 0.15 |
ENST00000537142.1
ENST00000448462.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr6_+_30689350 | 0.15 |
ENST00000330914.3
|
TUBB
|
tubulin, beta class I |
| chr15_+_91446961 | 0.15 |
ENST00000559965.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr19_+_13906250 | 0.14 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr1_-_9189144 | 0.14 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr6_+_30689401 | 0.14 |
ENST00000396389.1
ENST00000396384.1 |
TUBB
|
tubulin, beta class I |
| chr20_+_44035200 | 0.13 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr16_-_402639 | 0.13 |
ENST00000262320.3
|
AXIN1
|
axin 1 |
| chr9_+_34989638 | 0.13 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr14_-_69446034 | 0.13 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr19_+_50180507 | 0.12 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr3_+_10206545 | 0.12 |
ENST00000256458.4
|
IRAK2
|
interleukin-1 receptor-associated kinase 2 |
| chr11_-_507184 | 0.12 |
ENST00000533410.1
ENST00000354420.2 ENST00000397604.3 ENST00000531149.1 ENST00000356187.5 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr14_+_64971438 | 0.12 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr19_+_10400615 | 0.12 |
ENST00000221980.4
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin |
| chr11_-_125365435 | 0.12 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr12_-_1703331 | 0.12 |
ENST00000339235.3
|
FBXL14
|
F-box and leucine-rich repeat protein 14 |
| chr17_-_4890919 | 0.12 |
ENST00000572543.1
ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr1_+_155051379 | 0.12 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr1_-_94312706 | 0.12 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr6_-_30712313 | 0.11 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr22_+_37956479 | 0.11 |
ENST00000430687.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr19_+_47778119 | 0.11 |
ENST00000552360.2
|
PRR24
|
proline rich 24 |
| chr20_+_35202909 | 0.11 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr19_-_10530784 | 0.11 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr19_+_41882466 | 0.11 |
ENST00000436170.2
|
TMEM91
|
transmembrane protein 91 |
| chr19_+_45504688 | 0.11 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr4_+_8201091 | 0.11 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr11_-_64545941 | 0.11 |
ENST00000377387.1
|
SF1
|
splicing factor 1 |
| chr20_+_34894247 | 0.11 |
ENST00000373913.3
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr15_+_39873268 | 0.10 |
ENST00000397591.2
ENST00000260356.5 |
THBS1
|
thrombospondin 1 |
| chr10_-_102790852 | 0.10 |
ENST00000470414.1
ENST00000370215.3 |
PDZD7
|
PDZ domain containing 7 |
| chr8_-_33424636 | 0.10 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr11_-_64546202 | 0.10 |
ENST00000377390.3
ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1
|
splicing factor 1 |
| chr11_+_64009072 | 0.10 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr19_+_50180317 | 0.10 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr11_-_125366089 | 0.10 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr6_-_31697255 | 0.10 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr19_-_663171 | 0.10 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr15_-_93199069 | 0.10 |
ENST00000327355.5
|
FAM174B
|
family with sequence similarity 174, member B |
| chr16_-_55866997 | 0.10 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr15_+_75494214 | 0.10 |
ENST00000394987.4
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr4_+_103422471 | 0.10 |
ENST00000226574.4
ENST00000394820.4 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr22_-_30722866 | 0.10 |
ENST00000403477.3
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr15_-_71146480 | 0.10 |
ENST00000299213.8
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr18_-_43684186 | 0.10 |
ENST00000590406.1
ENST00000282050.2 ENST00000590324.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr2_+_30454390 | 0.10 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr11_-_125366018 | 0.09 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr6_+_12012536 | 0.09 |
ENST00000379388.2
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chrX_-_19905703 | 0.09 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr9_-_35115836 | 0.09 |
ENST00000378566.1
ENST00000378554.2 ENST00000322813.5 |
FAM214B
|
family with sequence similarity 214, member B |
| chr13_-_111214015 | 0.09 |
ENST00000267328.3
|
RAB20
|
RAB20, member RAS oncogene family |
| chr6_-_26108355 | 0.09 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr6_-_31697563 | 0.09 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr19_+_797392 | 0.09 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr2_-_61108449 | 0.09 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr3_-_88108212 | 0.09 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr2_+_191513789 | 0.09 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr17_-_72772462 | 0.09 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_-_16682987 | 0.09 |
ENST00000431408.1
ENST00000436553.2 ENST00000595753.1 |
SLC35E1
|
solute carrier family 35, member E1 |
| chr3_-_52273098 | 0.09 |
ENST00000499914.2
ENST00000305533.5 ENST00000597542.1 |
TWF2
TLR9
|
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr11_-_65640071 | 0.09 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr19_+_8455200 | 0.09 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr1_-_33647267 | 0.08 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr9_+_131709966 | 0.08 |
ENST00000372577.2
|
NUP188
|
nucleoporin 188kDa |
| chr3_-_169487617 | 0.08 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr17_-_4890649 | 0.08 |
ENST00000361571.5
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr7_+_142985467 | 0.08 |
ENST00000392925.2
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr1_+_178994939 | 0.08 |
ENST00000440702.1
|
FAM20B
|
family with sequence similarity 20, member B |
| chr11_-_44972299 | 0.08 |
ENST00000528473.1
|
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr1_+_156698743 | 0.08 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr15_-_34502197 | 0.08 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr1_-_6321035 | 0.08 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr5_-_141030943 | 0.08 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr11_+_66624527 | 0.08 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr15_+_31619013 | 0.08 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr19_-_10946871 | 0.08 |
ENST00000589638.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr16_+_29818857 | 0.08 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_+_2285817 | 0.08 |
ENST00000564065.1
|
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr4_-_82393009 | 0.08 |
ENST00000436139.2
|
RASGEF1B
|
RasGEF domain family, member 1B |
| chr16_+_69139467 | 0.08 |
ENST00000569188.1
|
HAS3
|
hyaluronan synthase 3 |
| chr19_+_40871734 | 0.08 |
ENST00000359274.5
|
PLD3
|
phospholipase D family, member 3 |
| chr11_+_64685026 | 0.08 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr5_-_176037105 | 0.08 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr19_-_51568324 | 0.08 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr6_-_110501200 | 0.08 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr14_-_24610779 | 0.08 |
ENST00000560403.1
ENST00000419198.2 ENST00000216799.4 |
EMC9
|
ER membrane protein complex subunit 9 |
| chr18_+_77794446 | 0.08 |
ENST00000262197.7
|
RBFA
|
ribosome binding factor A (putative) |
| chr11_-_506739 | 0.08 |
ENST00000529306.1
ENST00000438658.2 ENST00000527485.1 ENST00000397615.2 ENST00000397614.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr4_-_109087872 | 0.07 |
ENST00000510624.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr5_+_172068232 | 0.07 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr20_+_60878005 | 0.07 |
ENST00000253003.2
|
ADRM1
|
adhesion regulating molecule 1 |
| chr4_+_103422499 | 0.07 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr2_-_152118276 | 0.07 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr20_-_3996165 | 0.07 |
ENST00000545616.2
ENST00000358395.6 |
RNF24
|
ring finger protein 24 |
| chr6_+_35310312 | 0.07 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr4_+_83351791 | 0.07 |
ENST00000509635.1
|
ENOPH1
|
enolase-phosphatase 1 |
| chr1_-_161147275 | 0.07 |
ENST00000319769.5
ENST00000367998.1 |
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr8_+_32405728 | 0.07 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr16_-_28621353 | 0.07 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr17_-_4710288 | 0.07 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr8_-_57123815 | 0.07 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr17_-_72772425 | 0.07 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_-_663277 | 0.07 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chr16_-_2185899 | 0.07 |
ENST00000262304.4
ENST00000423118.1 |
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr7_-_108096765 | 0.07 |
ENST00000379024.4
ENST00000351718.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr19_+_11457232 | 0.07 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chrX_-_47479246 | 0.07 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr18_+_3449695 | 0.07 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr15_-_40213080 | 0.07 |
ENST00000561100.1
|
GPR176
|
G protein-coupled receptor 176 |
| chr19_+_19303720 | 0.07 |
ENST00000392324.4
|
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr3_+_88108381 | 0.07 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr1_+_41157361 | 0.07 |
ENST00000427410.2
ENST00000447388.3 ENST00000425457.2 ENST00000453631.1 ENST00000456393.2 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr19_-_344786 | 0.07 |
ENST00000264819.4
|
MIER2
|
mesoderm induction early response 1, family member 2 |
| chrX_+_133930798 | 0.07 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr16_-_30134266 | 0.07 |
ENST00000484663.1
ENST00000478356.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr19_+_11071685 | 0.07 |
ENST00000541122.2
ENST00000589677.1 ENST00000444061.3 |
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr14_-_64971288 | 0.07 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr17_-_41623691 | 0.07 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr14_+_100842735 | 0.07 |
ENST00000554998.1
ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25
|
WD repeat domain 25 |
| chr17_-_1083078 | 0.07 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr19_-_14201507 | 0.07 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr1_-_160068645 | 0.07 |
ENST00000448417.1
|
IGSF8
|
immunoglobulin superfamily, member 8 |
| chr20_+_44034676 | 0.07 |
ENST00000372723.3
ENST00000372722.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr15_+_90792760 | 0.07 |
ENST00000339615.5
ENST00000438251.1 |
TTLL13
|
tubulin tyrosine ligase-like family, member 13 |
| chr3_-_138048653 | 0.07 |
ENST00000460099.1
|
NME9
|
NME/NM23 family member 9 |
| chr1_+_155051305 | 0.07 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr11_-_47447970 | 0.07 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr10_-_101380121 | 0.07 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr19_-_48673552 | 0.06 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr11_-_65640198 | 0.06 |
ENST00000528176.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr1_+_114522049 | 0.06 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr1_-_155947951 | 0.06 |
ENST00000313695.7
ENST00000497907.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr6_+_144471643 | 0.06 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr12_-_125473600 | 0.06 |
ENST00000308736.2
|
DHX37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr11_+_111473108 | 0.06 |
ENST00000304987.3
|
SIK2
|
salt-inducible kinase 2 |
| chr16_-_838329 | 0.06 |
ENST00000563560.1
ENST00000569601.1 ENST00000565809.1 ENST00000565377.1 ENST00000007264.2 ENST00000567114.1 |
RPUSD1
|
RNA pseudouridylate synthase domain containing 1 |
| chr17_-_41623716 | 0.06 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr1_-_38471156 | 0.06 |
ENST00000373016.3
|
FHL3
|
four and a half LIM domains 3 |
| chr16_+_75182376 | 0.06 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr19_-_13617247 | 0.06 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr19_-_55770311 | 0.06 |
ENST00000412770.2
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1 |
| chr19_-_13068012 | 0.06 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr12_+_49658855 | 0.06 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr20_-_61569227 | 0.06 |
ENST00000266070.4
ENST00000395335.2 ENST00000266071.5 |
DIDO1
|
death inducer-obliterator 1 |
| chr18_+_55102917 | 0.06 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr5_-_38845812 | 0.06 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr9_-_131709858 | 0.06 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr19_+_797443 | 0.06 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr18_-_43684230 | 0.06 |
ENST00000592989.1
ENST00000589869.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr7_-_108096822 | 0.06 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr1_+_156698708 | 0.06 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr19_+_7985880 | 0.06 |
ENST00000597584.1
|
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr22_+_31090793 | 0.06 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr14_+_65170820 | 0.06 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr5_-_162887054 | 0.06 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr15_-_75744014 | 0.06 |
ENST00000394947.3
ENST00000565264.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr16_+_3070313 | 0.06 |
ENST00000326577.4
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr1_-_38273840 | 0.06 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr20_-_17662705 | 0.06 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr22_+_29138013 | 0.06 |
ENST00000216027.3
ENST00000398941.2 |
HSCB
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr19_+_7968728 | 0.06 |
ENST00000397981.3
ENST00000545011.1 ENST00000397983.3 ENST00000397979.3 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chr14_+_75230011 | 0.06 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr16_-_74808710 | 0.06 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr6_-_85474219 | 0.06 |
ENST00000369663.5
|
TBX18
|
T-box 18 |
| chr1_-_155948218 | 0.06 |
ENST00000313667.4
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr6_-_31864977 | 0.06 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr1_-_160068465 | 0.06 |
ENST00000314485.7
ENST00000368086.1 |
IGSF8
|
immunoglobulin superfamily, member 8 |
| chr19_+_40854559 | 0.06 |
ENST00000598962.1
ENST00000409419.1 ENST00000409587.1 ENST00000602131.1 ENST00000409735.4 ENST00000600948.1 ENST00000356508.5 ENST00000596682.1 ENST00000594908.1 |
PLD3
|
phospholipase D family, member 3 |
| chr6_-_112575758 | 0.06 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr1_+_215256467 | 0.06 |
ENST00000391894.2
ENST00000444842.2 |
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr19_-_37096139 | 0.06 |
ENST00000585983.1
ENST00000585960.1 ENST00000586115.1 |
ZNF529
|
zinc finger protein 529 |
| chr6_+_35310391 | 0.06 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR3_EGR2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.4 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.3 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.2 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.2 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.1 | 0.2 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.1 | 0.4 | GO:0046985 | negative regulation of megakaryocyte differentiation(GO:0045653) positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:1902824 | cleavage furrow ingression(GO:0036090) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.2 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.1 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.0 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.0 | GO:0090340 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of pancreatic juice secretion(GO:0090187) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.0 | 0.0 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.0 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.0 | GO:0060129 | forebrain dorsal/ventral pattern formation(GO:0021798) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.0 | GO:0036517 | cardiac right atrium morphogenesis(GO:0003213) chemoattraction of serotonergic neuron axon(GO:0036517) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.0 | 0.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.0 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.0 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.0 | GO:0052552 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:1902339 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.0 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.0 | GO:0075341 | host cell PML body(GO:0075341) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.0 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.0 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.0 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |