Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
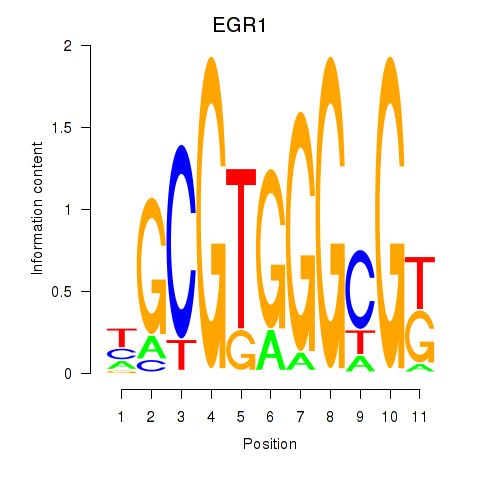
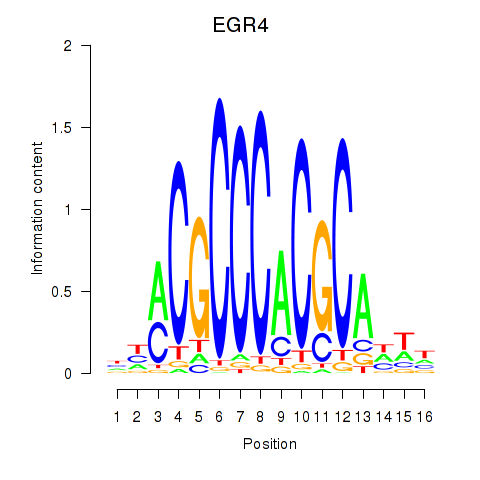
Results for EGR1_EGR4
Z-value: 0.77
Transcription factors associated with EGR1_EGR4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR1
|
ENSG00000120738.7 | early growth response 1 |
|
EGR4
|
ENSG00000135625.6 | early growth response 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR1 | hg19_v2_chr5_+_137801160_137801179 | -0.54 | 2.7e-01 | Click! |
Activity profile of EGR1_EGR4 motif
Sorted Z-values of EGR1_EGR4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_96928516 | 1.08 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr21_+_37529055 | 0.84 |
ENST00000270190.4
|
DOPEY2
|
dopey family member 2 |
| chr19_+_589893 | 0.53 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr7_-_44365020 | 0.46 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr1_+_35734616 | 0.44 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr15_-_80263506 | 0.41 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr12_+_105724613 | 0.39 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr2_+_86116396 | 0.35 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chrX_-_154033793 | 0.32 |
ENST00000369534.3
ENST00000413259.3 |
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr11_+_35965531 | 0.31 |
ENST00000528989.1
ENST00000524419.1 ENST00000315571.5 |
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3 |
| chr6_-_4135825 | 0.30 |
ENST00000380118.3
ENST00000413766.2 ENST00000361538.2 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chrX_-_154033686 | 0.28 |
ENST00000453245.1
ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr2_+_191513959 | 0.27 |
ENST00000337386.5
ENST00000357215.5 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr6_-_110500826 | 0.27 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr17_+_47439733 | 0.26 |
ENST00000507337.1
|
RP11-1079K10.3
|
RP11-1079K10.3 |
| chr12_+_105724414 | 0.26 |
ENST00000443585.1
ENST00000552457.1 ENST00000549893.1 |
C12orf75
|
chromosome 12 open reading frame 75 |
| chr12_+_130822606 | 0.26 |
ENST00000546060.1
ENST00000539400.1 |
PIWIL1
|
piwi-like RNA-mediated gene silencing 1 |
| chr19_-_39360561 | 0.24 |
ENST00000593809.1
ENST00000593424.1 |
RINL
|
Ras and Rab interactor-like |
| chr7_-_32931623 | 0.24 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr6_+_12012170 | 0.24 |
ENST00000487103.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr2_-_128145498 | 0.23 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr17_-_56065540 | 0.23 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr7_-_127032363 | 0.22 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr19_+_37096194 | 0.21 |
ENST00000460670.1
ENST00000292928.2 ENST00000439428.1 |
ZNF382
|
zinc finger protein 382 |
| chr1_-_38273840 | 0.21 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr2_+_99758161 | 0.21 |
ENST00000409684.1
|
C2ORF15
|
Uncharacterized protein C2orf15 |
| chr14_+_100070869 | 0.21 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr15_-_71146347 | 0.21 |
ENST00000559140.2
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr13_-_37494391 | 0.21 |
ENST00000379826.4
|
SMAD9
|
SMAD family member 9 |
| chr5_-_142784003 | 0.20 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr8_+_26149274 | 0.20 |
ENST00000522535.1
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr2_-_202316169 | 0.19 |
ENST00000430254.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr8_-_63951730 | 0.19 |
ENST00000260118.6
|
GGH
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chr7_-_100493744 | 0.19 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chrX_-_108976410 | 0.19 |
ENST00000504980.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr17_+_44668387 | 0.19 |
ENST00000576040.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr22_+_38597889 | 0.19 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr5_+_63461642 | 0.18 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chrX_-_108976521 | 0.18 |
ENST00000469796.2
ENST00000502391.1 ENST00000508092.1 ENST00000340800.2 ENST00000348502.6 |
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr6_-_4135693 | 0.18 |
ENST00000495548.1
ENST00000380125.2 ENST00000465828.1 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr9_+_129986734 | 0.18 |
ENST00000444677.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr2_+_191513789 | 0.18 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr16_+_23847339 | 0.18 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr1_+_65886262 | 0.18 |
ENST00000371065.4
|
LEPROT
|
leptin receptor overlapping transcript |
| chr12_+_107349606 | 0.17 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr5_-_145214848 | 0.17 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr8_-_102217515 | 0.17 |
ENST00000520347.1
ENST00000523922.1 ENST00000520984.1 |
ZNF706
|
zinc finger protein 706 |
| chr21_-_44527613 | 0.17 |
ENST00000380276.2
ENST00000398137.1 ENST00000291552.4 |
U2AF1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr1_+_212458834 | 0.17 |
ENST00000261461.2
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr10_-_75255724 | 0.17 |
ENST00000342558.3
ENST00000360663.5 ENST00000394829.2 ENST00000394828.2 ENST00000394822.2 |
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr15_+_52311398 | 0.17 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr21_+_17102311 | 0.16 |
ENST00000285679.6
ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25
|
ubiquitin specific peptidase 25 |
| chr12_-_64616019 | 0.16 |
ENST00000311915.8
ENST00000398055.3 ENST00000544871.1 |
C12orf66
|
chromosome 12 open reading frame 66 |
| chr5_-_145214893 | 0.16 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr1_+_6845578 | 0.16 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr19_-_49622348 | 0.16 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr16_-_31021717 | 0.16 |
ENST00000565419.1
|
STX1B
|
syntaxin 1B |
| chr9_+_91003271 | 0.16 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr12_-_109125285 | 0.15 |
ENST00000552871.1
ENST00000261401.3 |
CORO1C
|
coronin, actin binding protein, 1C |
| chr1_+_65886326 | 0.15 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chr16_+_23847267 | 0.15 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr3_+_37903432 | 0.15 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chrX_+_107068959 | 0.15 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr15_+_84116106 | 0.15 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr19_-_38714847 | 0.15 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr1_+_6052700 | 0.15 |
ENST00000378092.1
ENST00000445501.1 |
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr19_-_51568324 | 0.15 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr9_+_19230433 | 0.14 |
ENST00000434457.2
ENST00000602925.1 |
DENND4C
|
DENN/MADD domain containing 4C |
| chr22_-_29137771 | 0.14 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr13_+_76123883 | 0.14 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr16_+_29817841 | 0.14 |
ENST00000322945.6
ENST00000562337.1 ENST00000566906.2 ENST00000563402.1 ENST00000219782.6 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr17_+_46185111 | 0.14 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr13_-_37494365 | 0.14 |
ENST00000350148.5
|
SMAD9
|
SMAD family member 9 |
| chr1_+_6845497 | 0.14 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr18_-_51750948 | 0.14 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr9_+_114659046 | 0.14 |
ENST00000374279.3
|
UGCG
|
UDP-glucose ceramide glucosyltransferase |
| chr1_-_85156417 | 0.14 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr18_+_12948000 | 0.14 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr17_-_15466850 | 0.14 |
ENST00000438826.3
ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr7_+_33169142 | 0.14 |
ENST00000242067.6
ENST00000350941.3 ENST00000396127.2 ENST00000355070.2 ENST00000354265.4 ENST00000425508.2 |
BBS9
|
Bardet-Biedl syndrome 9 |
| chr14_-_51027838 | 0.14 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr14_-_53619816 | 0.14 |
ENST00000323669.5
ENST00000395606.1 ENST00000357758.3 |
DDHD1
|
DDHD domain containing 1 |
| chr6_+_138188378 | 0.13 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr9_-_13279589 | 0.13 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr4_+_26859300 | 0.13 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2 |
| chr7_-_35840198 | 0.13 |
ENST00000412856.1
ENST00000437235.3 ENST00000424194.1 |
AC007551.3
|
AC007551.3 |
| chr6_+_43739697 | 0.13 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr22_+_29469100 | 0.13 |
ENST00000327813.5
ENST00000407188.1 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr2_+_191513587 | 0.13 |
ENST00000416973.1
ENST00000426601.1 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr3_-_185542761 | 0.13 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr1_-_93426998 | 0.13 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr13_+_41635617 | 0.13 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr11_-_46142948 | 0.13 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr4_-_819901 | 0.13 |
ENST00000304062.6
|
CPLX1
|
complexin 1 |
| chr16_+_2588012 | 0.13 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr17_-_4269768 | 0.13 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr2_+_191745560 | 0.13 |
ENST00000338435.4
|
GLS
|
glutaminase |
| chr18_-_5295679 | 0.12 |
ENST00000582388.1
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr9_+_131451480 | 0.12 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chrX_-_107334790 | 0.12 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr5_-_137878887 | 0.12 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr1_+_65886244 | 0.12 |
ENST00000344610.8
|
LEPR
|
leptin receptor |
| chr19_+_52800410 | 0.12 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr3_+_196366616 | 0.12 |
ENST00000426755.1
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr20_+_31350184 | 0.12 |
ENST00000328111.2
ENST00000353855.2 ENST00000348286.2 |
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta |
| chr11_+_95523823 | 0.12 |
ENST00000538658.1
|
CEP57
|
centrosomal protein 57kDa |
| chrX_+_21959108 | 0.12 |
ENST00000457085.1
|
SMS
|
spermine synthase |
| chr9_-_86432547 | 0.12 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr4_-_185395191 | 0.12 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr18_+_22006646 | 0.12 |
ENST00000585067.1
ENST00000578221.1 |
IMPACT
|
impact RWD domain protein |
| chr7_+_43803790 | 0.12 |
ENST00000424330.1
|
BLVRA
|
biliverdin reductase A |
| chr7_+_100797726 | 0.12 |
ENST00000429457.1
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr12_-_96794330 | 0.12 |
ENST00000261211.3
|
CDK17
|
cyclin-dependent kinase 17 |
| chr12_+_12510045 | 0.12 |
ENST00000314565.4
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr14_-_45603657 | 0.12 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr13_-_52026730 | 0.12 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr5_+_172483347 | 0.12 |
ENST00000522692.1
ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF
|
CREB3 regulatory factor |
| chr21_+_38445539 | 0.12 |
ENST00000418766.1
ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3
|
tetratricopeptide repeat domain 3 |
| chr12_+_95867919 | 0.12 |
ENST00000261220.9
ENST00000549502.1 ENST00000553151.1 ENST00000550777.1 ENST00000551840.1 |
METAP2
|
methionyl aminopeptidase 2 |
| chr17_+_55334364 | 0.12 |
ENST00000322684.3
ENST00000579590.1 |
MSI2
|
musashi RNA-binding protein 2 |
| chr1_+_150898733 | 0.12 |
ENST00000525956.1
|
SETDB1
|
SET domain, bifurcated 1 |
| chr9_+_129987488 | 0.11 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr12_-_118498911 | 0.11 |
ENST00000544233.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr22_-_39548511 | 0.11 |
ENST00000434260.1
|
CBX7
|
chromobox homolog 7 |
| chr2_-_27341765 | 0.11 |
ENST00000405600.1
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr6_+_111195973 | 0.11 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr14_-_102553371 | 0.11 |
ENST00000553585.1
ENST00000216281.8 |
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr1_+_201924619 | 0.11 |
ENST00000367287.4
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr3_+_3168600 | 0.11 |
ENST00000251607.6
ENST00000339437.6 ENST00000280591.6 ENST00000420393.1 |
TRNT1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr15_+_86087267 | 0.11 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr5_+_179921344 | 0.11 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr16_+_21610879 | 0.11 |
ENST00000396014.4
|
METTL9
|
methyltransferase like 9 |
| chr18_-_658244 | 0.11 |
ENST00000585033.1
ENST00000323813.3 |
C18orf56
|
chromosome 18 open reading frame 56 |
| chr2_+_153191706 | 0.11 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr11_-_123525289 | 0.11 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr16_+_1203194 | 0.11 |
ENST00000348261.5
ENST00000358590.4 |
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr1_-_23694794 | 0.11 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr6_+_12012536 | 0.11 |
ENST00000379388.2
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr1_-_38471156 | 0.11 |
ENST00000373016.3
|
FHL3
|
four and a half LIM domains 3 |
| chr3_-_32612263 | 0.11 |
ENST00000432458.2
ENST00000424991.1 ENST00000273130.4 |
DYNC1LI1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr3_+_134514093 | 0.11 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr16_-_54962415 | 0.11 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chrX_-_101410762 | 0.11 |
ENST00000543160.1
ENST00000333643.3 |
BEX5
|
brain expressed, X-linked 5 |
| chr4_-_36246060 | 0.11 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr19_+_48112371 | 0.11 |
ENST00000594866.1
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1 |
| chr14_-_21945057 | 0.11 |
ENST00000397762.1
|
RAB2B
|
RAB2B, member RAS oncogene family |
| chr17_-_15466742 | 0.11 |
ENST00000584811.1
ENST00000419890.2 |
TVP23C
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr3_-_135915401 | 0.11 |
ENST00000491050.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr14_-_69658127 | 0.11 |
ENST00000556182.1
|
RP11-363J20.1
|
RP11-363J20.1 |
| chr21_+_44394742 | 0.11 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr11_+_102980126 | 0.11 |
ENST00000375735.2
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr1_-_197744763 | 0.11 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chrX_-_154033661 | 0.11 |
ENST00000393531.1
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr3_+_11196206 | 0.11 |
ENST00000431010.2
|
HRH1
|
histamine receptor H1 |
| chr2_-_98612379 | 0.10 |
ENST00000425805.2
|
TMEM131
|
transmembrane protein 131 |
| chr6_+_12008986 | 0.10 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr1_+_35734562 | 0.10 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chrX_+_135579238 | 0.10 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr3_-_185542817 | 0.10 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr14_+_23352374 | 0.10 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr5_+_179921430 | 0.10 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr2_+_30370382 | 0.10 |
ENST00000402708.1
|
YPEL5
|
yippee-like 5 (Drosophila) |
| chr3_+_75721428 | 0.10 |
ENST00000463183.1
|
LINC00960
|
long intergenic non-protein coding RNA 960 |
| chr17_+_36508826 | 0.10 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr5_-_54523143 | 0.10 |
ENST00000513312.1
|
MCIDAS
|
multiciliate differentiation and DNA synthesis associated cell cycle protein |
| chr10_+_98592009 | 0.10 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr2_+_64068074 | 0.10 |
ENST00000394417.2
ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr2_-_38829768 | 0.10 |
ENST00000378915.3
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr1_-_204119009 | 0.10 |
ENST00000444817.1
|
ETNK2
|
ethanolamine kinase 2 |
| chr10_-_95462265 | 0.10 |
ENST00000536233.1
ENST00000359204.4 ENST00000371430.2 ENST00000394100.2 |
FRA10AC1
|
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 |
| chr1_+_235490659 | 0.10 |
ENST00000488594.1
|
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr12_-_112856623 | 0.10 |
ENST00000551291.2
|
RPL6
|
ribosomal protein L6 |
| chr9_-_13279563 | 0.10 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr2_-_148778323 | 0.10 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr22_-_24181174 | 0.10 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr2_+_191208601 | 0.10 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr14_-_64971288 | 0.10 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr8_+_57124245 | 0.10 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr14_-_23822080 | 0.10 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr1_+_226250379 | 0.10 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr8_-_74791051 | 0.10 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr18_+_12947981 | 0.10 |
ENST00000262124.11
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr11_-_46638720 | 0.10 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr2_+_191208196 | 0.10 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr11_-_10315741 | 0.10 |
ENST00000256190.8
|
SBF2
|
SET binding factor 2 |
| chr9_+_96338647 | 0.10 |
ENST00000359246.4
|
PHF2
|
PHD finger protein 2 |
| chr12_-_49351148 | 0.10 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr22_+_19706958 | 0.10 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr1_+_51434357 | 0.10 |
ENST00000396148.1
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr15_-_65282232 | 0.10 |
ENST00000416889.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr1_-_98386543 | 0.09 |
ENST00000423006.2
ENST00000370192.3 ENST00000306031.5 |
DPYD
|
dihydropyrimidine dehydrogenase |
| chr4_-_100867864 | 0.09 |
ENST00000442697.2
|
DNAJB14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr5_+_145583156 | 0.09 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr4_+_103790120 | 0.09 |
ENST00000273986.4
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr6_+_31543334 | 0.09 |
ENST00000449264.2
|
TNF
|
tumor necrosis factor |
| chr11_+_95523621 | 0.09 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr15_-_65282274 | 0.09 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr21_+_34602377 | 0.09 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr8_-_144679602 | 0.09 |
ENST00000526710.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr19_-_50836762 | 0.09 |
ENST00000474951.1
ENST00000391818.2 |
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR1_EGR4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.2 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.2 | GO:0060733 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.1 | 0.1 | GO:2000049 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.5 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.1 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.4 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.7 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:2000672 | cellular response to sorbitol(GO:0072709) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0014744 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of muscle adaptation(GO:0014744) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.0 | GO:0061441 | renal artery morphogenesis(GO:0061441) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0030862 | positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.0 | 0.0 | GO:0033319 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.0 | 0.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.0 | GO:0098700 | sequestering of neurotransmitter(GO:0042137) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:0060129 | forebrain dorsal/ventral pattern formation(GO:0021798) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.0 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.0 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.0 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:2000230 | pancreatic stellate cell proliferation(GO:0072343) response to metformin(GO:1901558) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.0 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0071389 | cellular response to mineralocorticoid stimulus(GO:0071389) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.3 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.5 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.3 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.2 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.3 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.2 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.0 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.0 | 0.1 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.0 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.0 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |