Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
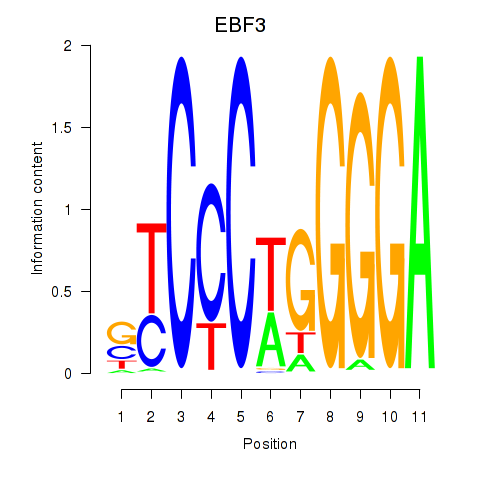
Results for EBF3
Z-value: 1.04
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.9 | EBF transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EBF3 | hg19_v2_chr10_-_131762105_131762105 | -0.15 | 7.8e-01 | Click! |
Activity profile of EBF3 motif
Sorted Z-values of EBF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_1051455 | 1.22 |
ENST00000379339.1
ENST00000480643.1 ENST00000434641.1 ENST00000421241.2 |
C1orf159
|
chromosome 1 open reading frame 159 |
| chr14_-_89960395 | 0.88 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr18_-_19748379 | 0.73 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr17_-_65992544 | 0.71 |
ENST00000580729.1
|
RP11-855A2.5
|
RP11-855A2.5 |
| chr12_-_49259643 | 0.68 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr10_+_77056181 | 0.55 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr17_-_6616678 | 0.55 |
ENST00000381074.4
ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr1_+_153330322 | 0.53 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr12_+_7072354 | 0.51 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr14_+_24630465 | 0.49 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr16_+_67233007 | 0.45 |
ENST00000360833.1
ENST00000393997.2 |
ELMO3
|
engulfment and cell motility 3 |
| chr15_+_96897466 | 0.44 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr17_+_77021702 | 0.44 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr3_+_32280159 | 0.43 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr20_-_44516256 | 0.42 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr19_-_42192189 | 0.42 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr14_-_69350920 | 0.42 |
ENST00000553290.1
|
ACTN1
|
actinin, alpha 1 |
| chr1_+_895930 | 0.40 |
ENST00000338591.3
|
KLHL17
|
kelch-like family member 17 |
| chr6_-_41908428 | 0.40 |
ENST00000505064.1
|
CCND3
|
cyclin D3 |
| chr5_-_180018540 | 0.39 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr11_+_124735282 | 0.39 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr5_+_3596168 | 0.39 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr6_+_292253 | 0.39 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr16_+_30662085 | 0.39 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr9_+_140317802 | 0.37 |
ENST00000341349.2
ENST00000392815.2 |
NOXA1
|
NADPH oxidase activator 1 |
| chr20_-_43743790 | 0.37 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr19_+_1495362 | 0.36 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr8_-_25281747 | 0.36 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr19_+_50180409 | 0.36 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr19_+_54369434 | 0.35 |
ENST00000421337.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr17_-_80023401 | 0.34 |
ENST00000354321.7
ENST00000306796.5 |
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr11_-_58275578 | 0.34 |
ENST00000360374.2
|
OR5B21
|
olfactory receptor, family 5, subfamily B, member 21 |
| chr10_+_99332198 | 0.34 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr9_-_139581848 | 0.34 |
ENST00000538402.1
ENST00000371694.3 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr11_-_9482010 | 0.33 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr17_+_77020224 | 0.33 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_+_984313 | 0.33 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr12_-_106697974 | 0.33 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr17_+_73629500 | 0.32 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr17_+_77020325 | 0.32 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr16_-_30798492 | 0.32 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr9_-_139581875 | 0.31 |
ENST00000371696.2
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr19_+_55996565 | 0.31 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr17_-_73874654 | 0.31 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr14_+_105953246 | 0.31 |
ENST00000392531.3
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr15_-_40633101 | 0.31 |
ENST00000559313.1
|
C15orf52
|
chromosome 15 open reading frame 52 |
| chr16_+_81772633 | 0.31 |
ENST00000566191.1
ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1
PLCG2
|
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr3_-_161089289 | 0.30 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr22_-_19466643 | 0.29 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr16_+_67233412 | 0.28 |
ENST00000477898.1
|
ELMO3
|
engulfment and cell motility 3 |
| chr4_+_48988259 | 0.28 |
ENST00000226432.4
|
CWH43
|
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) |
| chr11_-_8739383 | 0.28 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr14_+_105953204 | 0.27 |
ENST00000409393.2
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr2_-_97523721 | 0.27 |
ENST00000393537.4
|
ANKRD39
|
ankyrin repeat domain 39 |
| chr11_+_118826999 | 0.26 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr16_+_89988259 | 0.26 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr12_-_112546547 | 0.26 |
ENST00000547133.1
|
NAA25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
| chr20_-_62203808 | 0.26 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr5_+_159656437 | 0.26 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr11_-_67120974 | 0.26 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr16_+_2083265 | 0.26 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr12_-_48152853 | 0.26 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr8_+_22446763 | 0.26 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr19_+_33668509 | 0.26 |
ENST00000592484.1
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr16_+_30671223 | 0.26 |
ENST00000568722.1
|
FBRS
|
fibrosin |
| chr19_+_50180507 | 0.25 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr1_+_110453203 | 0.25 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_+_3388181 | 0.25 |
ENST00000418137.1
ENST00000413250.2 |
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr6_-_32191834 | 0.25 |
ENST00000375023.3
|
NOTCH4
|
notch 4 |
| chr11_+_65480222 | 0.25 |
ENST00000534681.1
|
KAT5
|
K(lysine) acetyltransferase 5 |
| chr15_-_86338134 | 0.25 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr19_+_18043810 | 0.25 |
ENST00000445755.2
|
CCDC124
|
coiled-coil domain containing 124 |
| chr19_-_4722705 | 0.24 |
ENST00000598360.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr1_+_14925173 | 0.24 |
ENST00000376030.2
ENST00000503743.1 ENST00000422387.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr9_+_136325089 | 0.24 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr3_+_42695176 | 0.24 |
ENST00000232974.6
ENST00000457842.3 |
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr16_+_3068393 | 0.24 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr2_+_120189422 | 0.24 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr5_-_175964366 | 0.24 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr15_+_63188009 | 0.24 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr15_-_86338100 | 0.23 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr14_-_94595993 | 0.23 |
ENST00000238609.3
|
IFI27L2
|
interferon, alpha-inducible protein 27-like 2 |
| chr1_-_27693349 | 0.23 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chrX_+_48367338 | 0.23 |
ENST00000359882.4
ENST00000537758.1 ENST00000367574.4 ENST00000355961.4 ENST00000489940.1 ENST00000361988.3 |
PORCN
|
porcupine homolog (Drosophila) |
| chr14_+_105952648 | 0.23 |
ENST00000330233.7
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr8_-_19459993 | 0.23 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr4_+_39408470 | 0.23 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr22_-_19466454 | 0.22 |
ENST00000494054.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr19_+_49128209 | 0.22 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr1_-_44497024 | 0.22 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr17_+_77020146 | 0.22 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_75315654 | 0.22 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr8_+_145582217 | 0.22 |
ENST00000530047.1
ENST00000527078.1 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr17_-_27053216 | 0.22 |
ENST00000292090.3
|
TLCD1
|
TLC domain containing 1 |
| chr17_-_78194147 | 0.21 |
ENST00000534910.1
ENST00000326317.6 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr16_+_30662184 | 0.21 |
ENST00000300835.4
|
PRR14
|
proline rich 14 |
| chr19_-_40324255 | 0.21 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr8_-_145641864 | 0.21 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr14_+_55034599 | 0.21 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr1_-_11751665 | 0.21 |
ENST00000376667.3
ENST00000235310.3 |
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr11_-_65381643 | 0.21 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr16_+_30662360 | 0.21 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chr1_+_110453514 | 0.20 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr17_-_80023659 | 0.20 |
ENST00000578907.1
ENST00000577907.1 ENST00000578176.1 ENST00000582529.1 |
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr1_-_159924006 | 0.20 |
ENST00000368092.3
ENST00000368093.3 |
SLAMF9
|
SLAM family member 9 |
| chr11_+_20044375 | 0.20 |
ENST00000525322.1
ENST00000530408.1 |
NAV2
|
neuron navigator 2 |
| chr11_-_132813566 | 0.20 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr5_-_142065612 | 0.20 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr17_-_79792909 | 0.20 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr2_+_132044330 | 0.20 |
ENST00000416266.1
|
CYP4F31P
|
cytochrome P450, family 4, subfamily F, polypeptide 31, pseudogene |
| chr8_+_145582633 | 0.20 |
ENST00000540505.1
|
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr12_+_113860160 | 0.20 |
ENST00000553248.1
ENST00000345635.4 ENST00000547802.1 |
SDSL
|
serine dehydratase-like |
| chr7_-_100183742 | 0.19 |
ENST00000310300.6
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr19_+_39904168 | 0.19 |
ENST00000438123.1
ENST00000409797.2 ENST00000451354.2 |
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr19_+_7600584 | 0.19 |
ENST00000600737.1
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr19_+_50270219 | 0.19 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr7_+_3340989 | 0.19 |
ENST00000404826.2
ENST00000389531.3 |
SDK1
|
sidekick cell adhesion molecule 1 |
| chr15_+_66994561 | 0.18 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr19_+_50180317 | 0.18 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr11_-_10920714 | 0.18 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr19_+_50706866 | 0.18 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr9_-_138987115 | 0.18 |
ENST00000277554.2
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing |
| chr16_-_3068171 | 0.18 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr11_+_114128522 | 0.18 |
ENST00000535401.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr10_-_135171510 | 0.17 |
ENST00000278025.4
ENST00000368552.3 |
FUOM
|
fucose mutarotase |
| chr3_+_239652 | 0.17 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr22_+_37257015 | 0.17 |
ENST00000447071.1
ENST00000248899.6 ENST00000397147.4 |
NCF4
|
neutrophil cytosolic factor 4, 40kDa |
| chr8_+_145582231 | 0.17 |
ENST00000526338.1
ENST00000402965.1 ENST00000534725.1 ENST00000532887.1 ENST00000329994.2 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr19_-_1650666 | 0.17 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr11_+_75273101 | 0.17 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr1_-_26197744 | 0.17 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr1_-_154943212 | 0.17 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr19_-_18197799 | 0.17 |
ENST00000430026.3
ENST00000593993.2 |
IL12RB1
|
interleukin 12 receptor, beta 1 |
| chr19_-_59010565 | 0.17 |
ENST00000594786.1
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr11_-_7961141 | 0.17 |
ENST00000360759.3
|
OR10A3
|
olfactory receptor, family 10, subfamily A, member 3 |
| chr12_-_48152611 | 0.17 |
ENST00000389212.3
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr19_-_4722780 | 0.17 |
ENST00000600621.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr11_+_65408273 | 0.17 |
ENST00000394227.3
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr1_-_21606013 | 0.17 |
ENST00000357071.4
|
ECE1
|
endothelin converting enzyme 1 |
| chr9_+_35658262 | 0.16 |
ENST00000378407.3
ENST00000378406.1 ENST00000426546.2 ENST00000327351.2 ENST00000421582.2 |
CCDC107
|
coiled-coil domain containing 107 |
| chr5_-_172198190 | 0.16 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr16_+_71392616 | 0.16 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr17_+_73452545 | 0.16 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr11_-_10920838 | 0.16 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr22_-_19466683 | 0.16 |
ENST00000399523.1
ENST00000421968.2 ENST00000447868.1 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr1_+_2487631 | 0.16 |
ENST00000409119.1
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr19_+_2476116 | 0.16 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr1_+_209757051 | 0.16 |
ENST00000009105.1
ENST00000423146.1 ENST00000361322.2 |
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG |
| chr6_+_43738444 | 0.16 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr14_+_71788096 | 0.16 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_-_10334617 | 0.16 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.8 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr11_-_61124776 | 0.16 |
ENST00000542361.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr7_-_158380371 | 0.16 |
ENST00000389418.4
ENST00000389416.4 |
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr19_+_55795493 | 0.16 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr7_-_73133959 | 0.15 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr4_+_7194247 | 0.15 |
ENST00000507866.2
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr17_-_61776522 | 0.15 |
ENST00000582055.1
|
LIMD2
|
LIM domain containing 2 |
| chr5_+_176560595 | 0.15 |
ENST00000508896.1
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr19_-_7698599 | 0.15 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr1_-_1310530 | 0.15 |
ENST00000338370.3
ENST00000321751.5 ENST00000378853.3 |
AURKAIP1
|
aurora kinase A interacting protein 1 |
| chr22_-_19466732 | 0.15 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr2_-_238499131 | 0.15 |
ENST00000538644.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr5_+_140625147 | 0.15 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr15_+_90728145 | 0.15 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr16_+_72142195 | 0.15 |
ENST00000563819.1
ENST00000567142.2 |
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr8_-_6735451 | 0.15 |
ENST00000297439.3
|
DEFB1
|
defensin, beta 1 |
| chr16_+_30662050 | 0.15 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr3_-_49170405 | 0.15 |
ENST00000305544.4
ENST00000494831.1 |
LAMB2
|
laminin, beta 2 (laminin S) |
| chr17_-_42994283 | 0.15 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr17_+_7308172 | 0.15 |
ENST00000575301.1
|
NLGN2
|
neuroligin 2 |
| chr17_-_1395954 | 0.15 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr10_+_11784360 | 0.14 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr15_+_90544532 | 0.14 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr19_-_38743878 | 0.14 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr14_-_68000442 | 0.14 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr3_-_119379427 | 0.14 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr11_-_64545941 | 0.14 |
ENST00000377387.1
|
SF1
|
splicing factor 1 |
| chr16_-_89007491 | 0.14 |
ENST00000327483.5
ENST00000564416.1 |
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr19_+_4229495 | 0.14 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr3_+_173116225 | 0.14 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr20_+_62496596 | 0.14 |
ENST00000369927.4
ENST00000346249.4 ENST00000348257.5 ENST00000352482.4 ENST00000351424.4 ENST00000217121.5 ENST00000358548.4 |
TPD52L2
|
tumor protein D52-like 2 |
| chr22_+_29702572 | 0.14 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr10_-_135171479 | 0.13 |
ENST00000447176.1
|
FUOM
|
fucose mutarotase |
| chr3_+_159481464 | 0.13 |
ENST00000467377.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr4_-_5890145 | 0.13 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr11_-_72414430 | 0.13 |
ENST00000452383.2
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr10_+_104678032 | 0.13 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chr11_-_72433346 | 0.13 |
ENST00000334211.8
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr19_+_41107249 | 0.13 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr14_-_105635090 | 0.13 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr6_+_43737939 | 0.13 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr22_+_18560743 | 0.13 |
ENST00000399744.3
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr16_-_67867749 | 0.13 |
ENST00000566758.1
ENST00000445712.2 ENST00000219172.3 ENST00000564817.1 |
CENPT
|
centromere protein T |
| chr14_+_55034330 | 0.13 |
ENST00000251091.5
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr16_-_65155833 | 0.13 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr3_+_150804676 | 0.13 |
ENST00000474524.1
ENST00000273432.4 |
MED12L
|
mediator complex subunit 12-like |
| chr12_-_48152428 | 0.13 |
ENST00000449771.2
ENST00000395358.3 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr13_-_52980263 | 0.13 |
ENST00000258613.4
ENST00000544466.1 |
THSD1
|
thrombospondin, type I, domain containing 1 |
| chr16_-_29478016 | 0.12 |
ENST00000549858.1
ENST00000551411.1 |
RP11-345J4.3
|
Uncharacterized protein |
| chr1_-_221509638 | 0.12 |
ENST00000439004.1
|
RP11-421L10.1
|
RP11-421L10.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 1.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 0.8 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.6 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.8 | GO:0046985 | negative regulation of megakaryocyte differentiation(GO:0045653) positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.6 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.4 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.3 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.4 | GO:1904139 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.3 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.1 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.2 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.4 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.3 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.2 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.5 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:1904000 | positive regulation of growth rate(GO:0040010) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) positive regulation of eating behavior(GO:1904000) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.6 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.0 | 0.4 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.5 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:1904833 | negative regulation of sperm motility(GO:1901318) positive regulation of superoxide dismutase activity(GO:1901671) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:1904714 | chaperone-mediated autophagy(GO:0061684) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.3 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0021557 | optic cup structural organization(GO:0003409) oculomotor nerve development(GO:0021557) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.3 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.0 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.0 | GO:1900168 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.0 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0051493 | regulation of cytoskeleton organization(GO:0051493) |
| 0.0 | 0.1 | GO:2000510 | peptide antigen assembly with MHC protein complex(GO:0002501) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.6 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.5 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.8 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.3 | GO:0016250 | N-sulfoglucosamine sulfohydrolase activity(GO:0016250) hydrolase activity, acting on acid sulfur-nitrogen bonds(GO:0016826) |
| 0.1 | 0.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.2 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.2 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 0.1 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |