Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
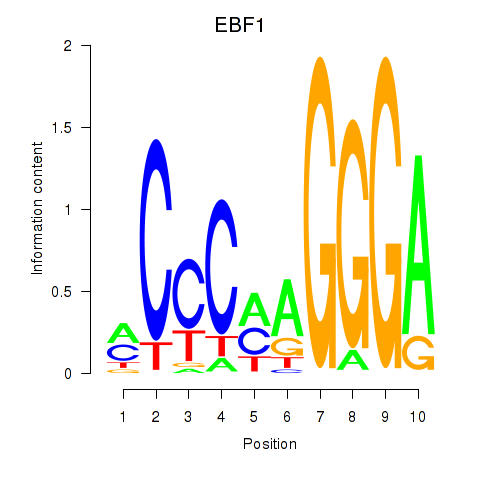
Results for EBF1
Z-value: 1.48
Transcription factors associated with EBF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF1
|
ENSG00000164330.12 | EBF transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EBF1 | hg19_v2_chr5_-_158526756_158526797 | -0.86 | 3.0e-02 | Click! |
Activity profile of EBF1 motif
Sorted Z-values of EBF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_27998689 | 1.37 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr14_+_103592636 | 1.28 |
ENST00000333007.1
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr12_-_49259643 | 1.28 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr2_-_241835561 | 1.00 |
ENST00000388934.4
|
C2orf54
|
chromosome 2 open reading frame 54 |
| chr19_-_50990785 | 0.95 |
ENST00000595005.1
|
CTD-2545M3.8
|
CTD-2545M3.8 |
| chr1_-_47655686 | 0.93 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr6_+_31895467 | 0.92 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr6_-_31550192 | 0.90 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr6_+_31895480 | 0.79 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr15_-_74494779 | 0.78 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr1_-_1051455 | 0.76 |
ENST00000379339.1
ENST00000480643.1 ENST00000434641.1 ENST00000421241.2 |
C1orf159
|
chromosome 1 open reading frame 159 |
| chr12_-_108991778 | 0.72 |
ENST00000549447.1
|
TMEM119
|
transmembrane protein 119 |
| chr11_-_795400 | 0.71 |
ENST00000526152.1
ENST00000456706.2 ENST00000528936.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr17_-_73874654 | 0.67 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr16_-_75467274 | 0.66 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr17_+_37821593 | 0.65 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr10_+_88718397 | 0.65 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr14_+_24630465 | 0.63 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr14_-_69350920 | 0.62 |
ENST00000553290.1
|
ACTN1
|
actinin, alpha 1 |
| chr17_-_4689649 | 0.62 |
ENST00000441199.2
ENST00000416307.2 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr2_+_74212073 | 0.61 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr18_-_19748379 | 0.58 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr1_+_161068179 | 0.57 |
ENST00000368011.4
ENST00000392192.2 |
KLHDC9
|
kelch domain containing 9 |
| chr19_+_33668509 | 0.54 |
ENST00000592484.1
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr5_+_3596168 | 0.53 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr5_+_148443049 | 0.52 |
ENST00000515304.1
ENST00000507318.1 |
CTC-529P8.1
|
CTC-529P8.1 |
| chr19_+_46498704 | 0.52 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr15_-_74495188 | 0.51 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr17_+_6916957 | 0.51 |
ENST00000547302.2
|
RNASEK-C17orf49
|
RNASEK-C17orf49 readthrough |
| chr1_-_45805752 | 0.51 |
ENST00000354383.6
ENST00000355498.2 ENST00000372100.5 ENST00000531105.1 |
MUTYH
|
mutY homolog |
| chr19_+_17326141 | 0.51 |
ENST00000445667.2
ENST00000263897.5 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr16_+_81772633 | 0.51 |
ENST00000566191.1
ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1
PLCG2
|
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr11_+_65686952 | 0.50 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr16_-_67260691 | 0.50 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr11_-_9482010 | 0.49 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr6_+_30851205 | 0.49 |
ENST00000515881.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr9_+_19408919 | 0.49 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr17_+_7533439 | 0.48 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr19_+_49622646 | 0.48 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr7_-_100881109 | 0.48 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr3_+_32280159 | 0.48 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr11_-_795286 | 0.48 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr6_-_160147925 | 0.47 |
ENST00000535561.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr1_-_44497024 | 0.47 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr6_+_131571535 | 0.46 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr16_-_31076332 | 0.46 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr14_+_95078714 | 0.46 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr4_+_74702214 | 0.45 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr17_+_75315654 | 0.45 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr12_-_53242770 | 0.44 |
ENST00000304620.4
ENST00000547110.1 |
KRT78
|
keratin 78 |
| chr11_+_844067 | 0.44 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr16_-_2168079 | 0.44 |
ENST00000488185.2
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr11_+_65337901 | 0.44 |
ENST00000309328.3
ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1 |
| chr20_+_44637526 | 0.43 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr6_-_34524049 | 0.43 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr22_+_18893816 | 0.43 |
ENST00000608842.1
|
DGCR6
|
DiGeorge syndrome critical region gene 6 |
| chr16_+_30662360 | 0.43 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chr11_-_1776176 | 0.42 |
ENST00000429746.1
|
CTSD
|
cathepsin D |
| chr15_+_91446961 | 0.42 |
ENST00000559965.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr19_+_41107249 | 0.42 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr11_-_57089774 | 0.41 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr19_-_4182497 | 0.41 |
ENST00000597896.1
|
SIRT6
|
sirtuin 6 |
| chr6_+_32812568 | 0.40 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_+_12902289 | 0.40 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr16_+_30662085 | 0.40 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr1_-_153363452 | 0.40 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr19_+_984313 | 0.40 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr17_+_14277419 | 0.39 |
ENST00000436469.1
|
AC022816.2
|
AC022816.2 |
| chr8_+_145582231 | 0.39 |
ENST00000526338.1
ENST00000402965.1 ENST00000534725.1 ENST00000532887.1 ENST00000329994.2 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr17_+_78389247 | 0.39 |
ENST00000520136.2
ENST00000520284.1 ENST00000517795.1 ENST00000523228.1 ENST00000523828.1 ENST00000522200.1 ENST00000521565.1 ENST00000518907.1 ENST00000518644.1 ENST00000518901.1 |
ENDOV
|
endonuclease V |
| chr3_-_107941209 | 0.39 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr22_-_21905120 | 0.39 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr16_+_2079501 | 0.38 |
ENST00000563587.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr7_+_76109827 | 0.38 |
ENST00000446820.2
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr1_-_6545502 | 0.38 |
ENST00000535355.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr11_+_60691924 | 0.38 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr19_+_35739782 | 0.38 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739897 | 0.38 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr9_-_33402506 | 0.38 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr6_-_30684898 | 0.38 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr14_-_89960395 | 0.37 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr11_+_66624527 | 0.37 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_-_4182530 | 0.37 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr19_+_35739631 | 0.37 |
ENST00000602003.1
ENST00000360798.3 ENST00000354900.3 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr8_+_55466915 | 0.37 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr19_+_7981030 | 0.36 |
ENST00000565886.1
|
TGFBR3L
|
transforming growth factor, beta receptor III-like |
| chr19_+_10654854 | 0.36 |
ENST00000586477.1
ENST00000586863.1 |
ATG4D
|
autophagy related 4D, cysteine peptidase |
| chr19_-_10230562 | 0.36 |
ENST00000587146.1
ENST00000588709.1 ENST00000253108.4 |
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr8_+_145582217 | 0.36 |
ENST00000530047.1
ENST00000527078.1 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr19_+_36132631 | 0.36 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr20_+_44462749 | 0.36 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr7_+_76139925 | 0.36 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B |
| chr20_-_36794938 | 0.35 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr13_+_36920569 | 0.35 |
ENST00000379848.2
|
SPG20OS
|
SPG20 opposite strand |
| chr20_+_55967129 | 0.35 |
ENST00000371219.2
|
RBM38
|
RNA binding motif protein 38 |
| chr20_-_62203808 | 0.35 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr19_+_7599128 | 0.35 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr1_+_35225339 | 0.35 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr3_-_49823941 | 0.35 |
ENST00000321599.4
ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1
|
inositol hexakisphosphate kinase 1 |
| chr14_+_105953246 | 0.35 |
ENST00000392531.3
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr20_-_60942326 | 0.34 |
ENST00000370677.3
ENST00000370692.3 |
LAMA5
|
laminin, alpha 5 |
| chr5_+_159656437 | 0.34 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr19_-_41934635 | 0.34 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr10_-_35930219 | 0.34 |
ENST00000374694.1
|
FZD8
|
frizzled family receptor 8 |
| chr6_+_33043703 | 0.34 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr1_-_45805607 | 0.34 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr19_+_17326521 | 0.34 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr16_-_67260901 | 0.34 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr1_-_45806053 | 0.33 |
ENST00000412971.1
ENST00000372098.3 ENST00000372110.3 ENST00000529984.1 ENST00000528332.2 ENST00000372115.3 ENST00000450313.1 |
MUTYH
|
mutY homolog |
| chr20_-_36794902 | 0.33 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr7_+_150026938 | 0.33 |
ENST00000343855.4
|
ZBED6CL
|
ZBED6 C-terminal like |
| chr7_+_150065879 | 0.33 |
ENST00000397281.2
ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1
ZNF775
|
replication initiator 1 zinc finger protein 775 |
| chr2_+_89952792 | 0.33 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr19_+_1041187 | 0.33 |
ENST00000531467.1
|
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr13_-_111214015 | 0.33 |
ENST00000267328.3
|
RAB20
|
RAB20, member RAS oncogene family |
| chr5_-_149792295 | 0.33 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr16_-_67978016 | 0.33 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr7_-_8302298 | 0.32 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr3_-_49726104 | 0.32 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_-_8739383 | 0.32 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr9_+_140172200 | 0.32 |
ENST00000357503.2
|
TOR4A
|
torsin family 4, member A |
| chr16_+_2083265 | 0.32 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr14_+_105953204 | 0.32 |
ENST00000409393.2
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr8_+_23386557 | 0.32 |
ENST00000523930.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr1_+_874649 | 0.31 |
ENST00000455979.1
|
SAMD11
|
sterile alpha motif domain containing 11 |
| chr1_+_156024552 | 0.31 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr9_-_139581848 | 0.31 |
ENST00000538402.1
ENST00000371694.3 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr11_+_313503 | 0.31 |
ENST00000528780.1
ENST00000328221.5 |
IFITM1
|
interferon induced transmembrane protein 1 |
| chr4_-_57522673 | 0.31 |
ENST00000381255.3
ENST00000317745.7 ENST00000555760.2 ENST00000556614.2 |
HOPX
|
HOP homeobox |
| chr22_+_31199037 | 0.31 |
ENST00000424224.1
|
OSBP2
|
oxysterol binding protein 2 |
| chr20_+_61436146 | 0.31 |
ENST00000290291.6
|
OGFR
|
opioid growth factor receptor |
| chr5_-_73936544 | 0.31 |
ENST00000509127.2
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr19_+_7598890 | 0.31 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr16_-_8955601 | 0.31 |
ENST00000569398.1
ENST00000568968.1 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr1_+_2407754 | 0.31 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chrX_-_48326764 | 0.31 |
ENST00000413668.1
ENST00000441948.1 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr19_+_12944722 | 0.30 |
ENST00000591495.1
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr15_+_74509530 | 0.30 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr8_+_23386305 | 0.30 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr17_+_78388959 | 0.30 |
ENST00000518137.1
ENST00000520367.1 ENST00000523999.1 ENST00000323854.5 ENST00000522751.1 |
ENDOV
|
endonuclease V |
| chr16_+_28874345 | 0.30 |
ENST00000566209.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr8_-_42065187 | 0.30 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chr3_-_48541537 | 0.30 |
ENST00000443308.2
ENST00000417841.1 |
SHISA5
|
shisa family member 5 |
| chr17_-_72864739 | 0.30 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr19_-_1650666 | 0.30 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr4_+_74735102 | 0.30 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr1_+_1550795 | 0.30 |
ENST00000520777.1
ENST00000357210.4 ENST00000360522.4 ENST00000378710.3 ENST00000355826.5 ENST00000518681.1 ENST00000505820.2 |
MIB2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr17_-_13505219 | 0.30 |
ENST00000284110.1
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr20_+_44462430 | 0.29 |
ENST00000462307.1
|
SNX21
|
sorting nexin family member 21 |
| chr19_+_35630926 | 0.29 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr7_-_139756791 | 0.29 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr19_+_15218180 | 0.29 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr19_-_6720686 | 0.29 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr14_-_24804269 | 0.29 |
ENST00000310677.4
ENST00000554068.2 ENST00000559167.1 ENST00000561138.1 |
ADCY4
|
adenylate cyclase 4 |
| chr10_+_106034637 | 0.29 |
ENST00000401888.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr19_+_40877583 | 0.29 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr16_+_30751953 | 0.29 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr7_+_76139833 | 0.29 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr17_+_73629500 | 0.29 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr8_-_145652336 | 0.29 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr2_-_241836298 | 0.29 |
ENST00000414499.1
|
C2orf54
|
chromosome 2 open reading frame 54 |
| chr16_-_29478016 | 0.29 |
ENST00000549858.1
ENST00000551411.1 |
RP11-345J4.3
|
Uncharacterized protein |
| chr17_-_80023659 | 0.29 |
ENST00000578907.1
ENST00000577907.1 ENST00000578176.1 ENST00000582529.1 |
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr19_-_42916499 | 0.29 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr17_-_73844722 | 0.29 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chr16_-_70472946 | 0.29 |
ENST00000342907.2
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr1_+_1370903 | 0.28 |
ENST00000338660.5
ENST00000404702.3 ENST00000476993.1 ENST00000471398.1 |
VWA1
|
von Willebrand factor A domain containing 1 |
| chr17_+_38083977 | 0.28 |
ENST00000578802.1
ENST00000578478.1 ENST00000582263.1 |
RP11-387H17.4
|
RP11-387H17.4 |
| chr5_-_139422654 | 0.28 |
ENST00000289409.4
ENST00000358522.3 ENST00000378238.4 ENST00000289422.7 ENST00000361474.1 ENST00000545385.1 ENST00000394770.1 ENST00000541337.1 |
NRG2
|
neuregulin 2 |
| chr17_-_18585131 | 0.28 |
ENST00000443457.1
ENST00000583002.1 |
ZNF286B
|
zinc finger protein 286B |
| chr19_+_17326191 | 0.28 |
ENST00000595101.1
ENST00000596136.1 ENST00000379776.4 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr19_-_36499521 | 0.28 |
ENST00000397428.3
ENST00000503121.1 ENST00000340477.5 ENST00000324444.3 ENST00000490730.1 |
SYNE4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr6_-_30712313 | 0.28 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr16_+_56672571 | 0.28 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr7_+_76101379 | 0.28 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr22_+_37678505 | 0.28 |
ENST00000402997.1
ENST00000405206.3 |
CYTH4
|
cytohesin 4 |
| chr4_+_25656920 | 0.28 |
ENST00000513204.1
|
SLC34A2
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 2 |
| chr17_+_43238438 | 0.28 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr6_+_30848829 | 0.28 |
ENST00000508317.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_+_1752372 | 0.27 |
ENST00000382349.4
|
ONECUT3
|
one cut homeobox 3 |
| chr7_+_65540853 | 0.27 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr19_+_18496957 | 0.27 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr9_+_130922537 | 0.27 |
ENST00000372994.1
|
C9orf16
|
chromosome 9 open reading frame 16 |
| chr19_+_3572925 | 0.27 |
ENST00000333651.6
ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr12_+_48099858 | 0.27 |
ENST00000547799.1
|
RP1-197B17.3
|
RP1-197B17.3 |
| chr19_-_9938480 | 0.27 |
ENST00000585379.1
|
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr16_+_30709530 | 0.27 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr1_-_206671061 | 0.27 |
ENST00000367119.1
|
C1orf147
|
chromosome 1 open reading frame 147 |
| chr17_-_79817091 | 0.27 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr16_+_30194916 | 0.27 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr19_+_35739597 | 0.27 |
ENST00000361790.3
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr6_-_43478239 | 0.27 |
ENST00000372441.1
|
LRRC73
|
leucine rich repeat containing 73 |
| chr14_+_23305783 | 0.27 |
ENST00000547279.1
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr10_+_77056181 | 0.27 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr22_-_50699972 | 0.26 |
ENST00000395778.3
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr20_-_62199427 | 0.26 |
ENST00000427522.2
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr12_-_66275350 | 0.26 |
ENST00000536648.1
|
RP11-366L20.2
|
Uncharacterized protein |
| chr1_+_24117627 | 0.26 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr16_-_30134266 | 0.26 |
ENST00000484663.1
ENST00000478356.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr19_+_48949030 | 0.26 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) histone H3-K9 deacetylation(GO:1990619) |
| 0.3 | 1.5 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.3 | 1.3 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 1.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 0.6 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 0.6 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 1.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 0.9 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 0.5 | GO:1901076 | positive regulation of engulfment of apoptotic cell(GO:1901076) |
| 0.2 | 0.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 0.7 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 0.5 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 1.3 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 1.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.6 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.1 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.4 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.5 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 1.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 | 0.5 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.1 | 0.4 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.7 | GO:0060481 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 2.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.6 | GO:0032796 | uropod organization(GO:0032796) |
| 0.1 | 0.1 | GO:0002709 | regulation of T cell mediated immunity(GO:0002709) |
| 0.1 | 0.3 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.3 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.1 | 0.5 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.2 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.1 | 0.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.1 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) |
| 0.1 | 1.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.4 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.1 | 0.2 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.1 | 0.5 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.3 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.1 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.2 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.1 | 0.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 1.7 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.3 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.4 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.5 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 0.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.5 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.7 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.3 | GO:1904139 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.2 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.2 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.2 | GO:0018012 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.1 | 0.5 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.4 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.1 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.1 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.2 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.2 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.2 | GO:0070649 | polar body extrusion after meiotic divisions(GO:0040038) establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.2 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.4 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.3 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 1.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.0 | GO:0035483 | gastric motility(GO:0035482) gastric emptying(GO:0035483) |
| 0.0 | 0.1 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.2 | GO:0072312 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) |
| 0.0 | 0.2 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.3 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:0003409 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.1 | GO:1905204 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.3 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.3 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 1.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0010171 | body morphogenesis(GO:0010171) |
| 0.0 | 0.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.2 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 0.3 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.2 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.4 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.5 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) positive regulation of sensory perception of pain(GO:1904058) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.4 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 1.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.9 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.2 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.2 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.2 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:1902566 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.2 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.4 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.0 | 0.5 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.7 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.2 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.0 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0051100 | negative regulation of binding(GO:0051100) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.0 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:1905245 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 1.7 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.0 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.4 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:1903772 | virus maturation(GO:0019075) regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.0 | GO:0060319 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0061051 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.3 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 1.4 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0070105 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0014806 | smooth muscle hyperplasia(GO:0014806) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.0 | GO:0021604 | cranial nerve structural organization(GO:0021604) |
| 0.0 | 0.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.2 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.2 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 | 0.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.0 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.3 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.7 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 1.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.5 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.1 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.5 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 0.2 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.2 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.4 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 1.0 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0036026 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 2.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.0 | 0.6 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:1902710 | GABA receptor complex(GO:1902710) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0032994 | protein-lipid complex(GO:0032994) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.3 | 0.8 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 0.9 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 0.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.4 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.5 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.4 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.1 | 1.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.5 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.5 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.3 | GO:0016731 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.3 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.2 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.2 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.2 | GO:0016250 | N-sulfoglucosamine sulfohydrolase activity(GO:0016250) hydrolase activity, acting on acid sulfur-nitrogen bonds(GO:0016826) |
| 0.1 | 0.5 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.2 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.2 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.1 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.2 | GO:0015226 | secondary active organic cation transmembrane transporter activity(GO:0008513) amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.3 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.2 | GO:0030545 | receptor regulator activity(GO:0030545) |
| 0.0 | 0.0 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.2 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.4 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 1.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.2 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.1 | GO:0047726 | iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.3 | GO:0015321 | sodium:phosphate symporter activity(GO:0005436) sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0050135 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.1 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0098519 | phosphoserine phosphatase activity(GO:0004647) nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 1.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.0 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.1 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 3.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |