Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
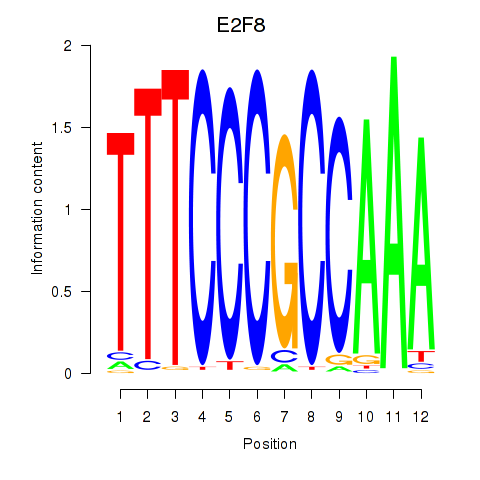
Results for E2F8
Z-value: 0.62
Transcription factors associated with E2F8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F8
|
ENSG00000129173.8 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F8 | hg19_v2_chr11_-_19263145_19263176 | -0.04 | 9.4e-01 | Click! |
Activity profile of E2F8 motif
Sorted Z-values of E2F8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_136633940 | 0.43 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr17_-_6915616 | 0.35 |
ENST00000575889.1
|
AC027763.2
|
Uncharacterized protein |
| chr8_-_25281747 | 0.31 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr8_-_120868078 | 0.28 |
ENST00000313655.4
|
DSCC1
|
DNA replication and sister chromatid cohesion 1 |
| chr19_+_57874835 | 0.25 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr2_+_24346324 | 0.25 |
ENST00000407625.1
ENST00000420135.2 |
FAM228B
|
family with sequence similarity 228, member B |
| chr4_+_1714548 | 0.25 |
ENST00000605571.1
|
RP11-572O17.1
|
RP11-572O17.1 |
| chr7_-_21985489 | 0.25 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr3_+_122296443 | 0.22 |
ENST00000464300.2
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr1_+_197871854 | 0.22 |
ENST00000436652.1
|
C1orf53
|
chromosome 1 open reading frame 53 |
| chr12_-_48499826 | 0.22 |
ENST00000551798.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr14_-_51135005 | 0.20 |
ENST00000556735.1
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chr9_-_8857776 | 0.18 |
ENST00000481079.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr19_+_37464440 | 0.18 |
ENST00000455817.1
ENST00000588596.1 |
ZNF568
|
zinc finger protein 568 |
| chr19_+_47760777 | 0.17 |
ENST00000599398.1
ENST00000595659.1 |
CCDC9
|
coiled-coil domain containing 9 |
| chr9_+_8858102 | 0.17 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr3_-_123168551 | 0.17 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr19_+_12203069 | 0.16 |
ENST00000430298.2
ENST00000339302.4 |
ZNF788
ZNF788
|
zinc finger family member 788 Zinc finger protein 788 |
| chr7_-_56118981 | 0.16 |
ENST00000419984.2
ENST00000413218.1 ENST00000424596.1 |
PSPH
|
phosphoserine phosphatase |
| chr19_-_51845378 | 0.16 |
ENST00000335624.4
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like |
| chr6_+_26204825 | 0.15 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr3_+_44754126 | 0.15 |
ENST00000449836.1
ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr22_+_19467261 | 0.15 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr12_+_107168418 | 0.14 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr15_-_100258029 | 0.14 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr5_+_172483347 | 0.14 |
ENST00000522692.1
ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF
|
CREB3 regulatory factor |
| chrX_-_53449593 | 0.14 |
ENST00000375340.6
ENST00000322213.4 |
SMC1A
|
structural maintenance of chromosomes 1A |
| chr4_-_174255400 | 0.14 |
ENST00000506267.1
|
HMGB2
|
high mobility group box 2 |
| chr5_-_132166303 | 0.14 |
ENST00000440118.1
|
SHROOM1
|
shroom family member 1 |
| chr17_-_15165854 | 0.13 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
| chr19_+_12203100 | 0.13 |
ENST00000596883.1
|
ZNF788
|
zinc finger family member 788 |
| chr17_+_38444115 | 0.13 |
ENST00000580824.1
ENST00000577249.1 |
CDC6
|
cell division cycle 6 |
| chr2_+_63816126 | 0.13 |
ENST00000454035.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr10_-_74385811 | 0.13 |
ENST00000603011.1
ENST00000401998.3 ENST00000361114.5 ENST00000604238.1 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr7_-_21985656 | 0.13 |
ENST00000406877.3
|
CDCA7L
|
cell division cycle associated 7-like |
| chr10_+_35416223 | 0.13 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr15_-_57025728 | 0.13 |
ENST00000559352.1
|
ZNF280D
|
zinc finger protein 280D |
| chr21_+_37477164 | 0.13 |
ENST00000422473.1
|
AP000688.29
|
AP000688.29 |
| chr11_+_47236489 | 0.13 |
ENST00000256996.4
ENST00000378603.3 ENST00000378600.3 ENST00000378601.3 |
DDB2
|
damage-specific DNA binding protein 2, 48kDa |
| chr3_+_14058794 | 0.13 |
ENST00000424053.1
ENST00000528067.1 ENST00000429201.1 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr6_+_135502501 | 0.13 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr18_-_47018897 | 0.12 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr7_+_128379449 | 0.12 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr12_-_8815299 | 0.12 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr1_+_7844312 | 0.12 |
ENST00000377541.1
|
PER3
|
period circadian clock 3 |
| chr3_+_14989186 | 0.12 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr10_-_17659234 | 0.12 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr9_+_114287433 | 0.12 |
ENST00000358151.4
ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483
|
zinc finger protein 483 |
| chr9_-_37904084 | 0.12 |
ENST00000377716.2
ENST00000242275.6 |
SLC25A51
|
solute carrier family 25, member 51 |
| chr3_+_181429704 | 0.11 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr1_+_48688357 | 0.11 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr8_+_118532937 | 0.11 |
ENST00000297347.3
|
MED30
|
mediator complex subunit 30 |
| chr7_-_152373216 | 0.11 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr10_-_46167722 | 0.11 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr2_+_10263298 | 0.11 |
ENST00000474701.1
|
RRM2
|
ribonucleotide reductase M2 |
| chr7_-_100026280 | 0.11 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr10_-_74385876 | 0.11 |
ENST00000398761.4
|
MICU1
|
mitochondrial calcium uptake 1 |
| chr17_-_9479128 | 0.11 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr7_+_116593433 | 0.11 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr3_-_107596910 | 0.11 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr2_-_113993020 | 0.11 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr12_-_107168696 | 0.11 |
ENST00000551505.1
|
RP11-144F15.1
|
Uncharacterized protein |
| chr11_+_125495862 | 0.11 |
ENST00000428830.2
ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1
|
checkpoint kinase 1 |
| chr6_-_85473156 | 0.11 |
ENST00000606784.1
ENST00000606325.1 |
TBX18
|
T-box 18 |
| chr20_-_45142154 | 0.11 |
ENST00000347606.4
ENST00000457685.2 |
ZNF334
|
zinc finger protein 334 |
| chr1_+_33283043 | 0.10 |
ENST00000373476.1
ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP
|
S100P binding protein |
| chr12_-_15942309 | 0.10 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chrX_-_135338503 | 0.10 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr17_+_61904766 | 0.10 |
ENST00000581842.1
ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5
|
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr19_-_58446721 | 0.10 |
ENST00000396147.1
ENST00000595569.1 ENST00000599852.1 ENST00000425570.3 ENST00000601593.1 |
ZNF418
|
zinc finger protein 418 |
| chr14_-_61190754 | 0.10 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr1_-_26232522 | 0.10 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr1_-_205719295 | 0.10 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr15_+_84116106 | 0.10 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr13_-_41345277 | 0.10 |
ENST00000323563.6
|
MRPS31
|
mitochondrial ribosomal protein S31 |
| chr18_-_47018869 | 0.10 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr1_-_110052302 | 0.10 |
ENST00000369864.4
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig-like domain 1 |
| chr4_-_39367949 | 0.10 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr4_-_174254823 | 0.10 |
ENST00000438704.2
|
HMGB2
|
high mobility group box 2 |
| chr7_+_116593292 | 0.10 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr8_+_69242957 | 0.09 |
ENST00000518698.1
ENST00000539993.1 |
C8orf34
|
chromosome 8 open reading frame 34 |
| chr12_+_111051902 | 0.09 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr16_-_103572 | 0.09 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr22_+_42086547 | 0.09 |
ENST00000402966.1
|
C22orf46
|
chromosome 22 open reading frame 46 |
| chr16_-_30134441 | 0.09 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr1_-_27709793 | 0.09 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr10_-_17659357 | 0.09 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr12_+_104682667 | 0.09 |
ENST00000527335.1
|
TXNRD1
|
thioredoxin reductase 1 |
| chr16_+_58074069 | 0.09 |
ENST00000570065.1
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr6_-_28973037 | 0.09 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr6_+_117002339 | 0.09 |
ENST00000413340.1
ENST00000368564.1 ENST00000356348.1 |
KPNA5
|
karyopherin alpha 5 (importin alpha 6) |
| chr3_-_125313934 | 0.09 |
ENST00000296220.5
|
OSBPL11
|
oxysterol binding protein-like 11 |
| chr12_-_123011476 | 0.09 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr6_+_135502466 | 0.09 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr16_-_18812746 | 0.09 |
ENST00000546206.2
ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1
RP11-1035H13.3
|
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr12_-_110939870 | 0.09 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr16_-_30134266 | 0.09 |
ENST00000484663.1
ENST00000478356.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr6_+_43603552 | 0.09 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr19_+_4402659 | 0.09 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr2_+_238395879 | 0.08 |
ENST00000445024.2
ENST00000338530.4 ENST00000409373.1 |
MLPH
|
melanophilin |
| chr18_+_21719018 | 0.08 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr15_+_84115868 | 0.08 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr1_+_43637996 | 0.08 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr7_-_56119156 | 0.08 |
ENST00000421312.1
ENST00000416592.1 |
PSPH
|
phosphoserine phosphatase |
| chr2_+_238395803 | 0.08 |
ENST00000264605.3
|
MLPH
|
melanophilin |
| chr11_-_107729287 | 0.08 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr16_-_82045049 | 0.08 |
ENST00000532128.1
ENST00000328945.5 |
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr1_-_43638168 | 0.08 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr6_-_143832793 | 0.08 |
ENST00000438118.2
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr1_+_172745006 | 0.08 |
ENST00000432694.2
|
RP1-15D23.2
|
RP1-15D23.2 |
| chr18_+_59992527 | 0.08 |
ENST00000586569.1
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr6_-_85473073 | 0.08 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr16_-_67185117 | 0.08 |
ENST00000449549.3
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr2_-_86790472 | 0.08 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chr17_-_60005329 | 0.08 |
ENST00000251334.6
|
INTS2
|
integrator complex subunit 2 |
| chr12_-_48499591 | 0.08 |
ENST00000551330.1
ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr18_+_21718924 | 0.08 |
ENST00000399496.3
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr17_+_66243715 | 0.08 |
ENST00000359904.3
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr1_+_224803995 | 0.08 |
ENST00000272133.3
|
CNIH3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr2_+_218989991 | 0.08 |
ENST00000453237.1
|
CXCR2
|
chemokine (C-X-C motif) receptor 2 |
| chr4_+_152330409 | 0.08 |
ENST00000513086.1
|
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr4_-_129209221 | 0.08 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr7_+_116593568 | 0.08 |
ENST00000446490.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr12_-_123011536 | 0.08 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr4_-_84406218 | 0.08 |
ENST00000515303.1
|
FAM175A
|
family with sequence similarity 175, member A |
| chr2_-_70780572 | 0.07 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr9_+_124088860 | 0.07 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr20_-_524340 | 0.07 |
ENST00000400227.3
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr12_-_8815215 | 0.07 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chrX_-_7895755 | 0.07 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr3_+_197677047 | 0.07 |
ENST00000448864.1
|
RPL35A
|
ribosomal protein L35a |
| chr11_+_9595180 | 0.07 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr11_-_107729504 | 0.07 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr5_-_132166579 | 0.07 |
ENST00000378679.3
|
SHROOM1
|
shroom family member 1 |
| chr15_+_44719394 | 0.07 |
ENST00000260327.4
ENST00000396780.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr8_+_71383312 | 0.07 |
ENST00000520259.1
|
RP11-333A23.4
|
RP11-333A23.4 |
| chr12_-_6772249 | 0.07 |
ENST00000467678.1
ENST00000493873.1 ENST00000423703.2 ENST00000412586.2 ENST00000444704.2 |
ING4
|
inhibitor of growth family, member 4 |
| chr22_-_30925150 | 0.07 |
ENST00000437871.1
|
SEC14L6
|
SEC14-like 6 (S. cerevisiae) |
| chr2_+_10262857 | 0.07 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr7_+_114562909 | 0.07 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr10_+_35416090 | 0.07 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr8_-_81083618 | 0.07 |
ENST00000520795.1
|
TPD52
|
tumor protein D52 |
| chr10_+_118083919 | 0.07 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr4_+_96012585 | 0.07 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr6_-_30899924 | 0.07 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr6_-_90348440 | 0.07 |
ENST00000520441.1
ENST00000520318.1 ENST00000523377.1 |
LYRM2
|
LYR motif containing 2 |
| chr1_-_43637915 | 0.07 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr12_+_111051832 | 0.06 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr15_-_57025675 | 0.06 |
ENST00000558320.1
|
ZNF280D
|
zinc finger protein 280D |
| chr14_-_23624511 | 0.06 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr10_-_43892279 | 0.06 |
ENST00000443950.2
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr12_-_6772303 | 0.06 |
ENST00000396807.4
ENST00000446105.2 ENST00000341550.4 |
ING4
|
inhibitor of growth family, member 4 |
| chr12_+_111843749 | 0.06 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr22_+_24105394 | 0.06 |
ENST00000305199.5
ENST00000382821.3 |
C22orf15
|
chromosome 22 open reading frame 15 |
| chr15_-_77712477 | 0.06 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr22_+_20104947 | 0.06 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr1_+_170501270 | 0.06 |
ENST00000367763.3
ENST00000367762.1 |
GORAB
|
golgin, RAB6-interacting |
| chr19_+_6740888 | 0.06 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr2_-_24583314 | 0.06 |
ENST00000443927.1
ENST00000406921.3 ENST00000412011.1 |
ITSN2
|
intersectin 2 |
| chr2_-_37193606 | 0.06 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr12_+_123011776 | 0.06 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr1_-_55230165 | 0.06 |
ENST00000371279.3
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr22_+_20105012 | 0.06 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr11_+_47586982 | 0.06 |
ENST00000426530.2
ENST00000534775.1 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr1_-_6295975 | 0.06 |
ENST00000343813.5
ENST00000362035.3 |
ICMT
|
isoprenylcysteine carboxyl methyltransferase |
| chr9_-_123639304 | 0.06 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr8_+_128426535 | 0.06 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr21_+_39644305 | 0.06 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr7_+_128379346 | 0.06 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chrX_+_131157322 | 0.06 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr14_-_61747949 | 0.06 |
ENST00000355702.2
|
TMEM30B
|
transmembrane protein 30B |
| chr2_-_230787879 | 0.06 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_-_57113314 | 0.06 |
ENST00000338458.4
ENST00000468727.1 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr3_-_25824925 | 0.06 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr10_-_12238071 | 0.06 |
ENST00000491614.1
ENST00000537776.1 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr8_-_131455835 | 0.06 |
ENST00000518721.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr15_+_44719970 | 0.06 |
ENST00000558966.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr11_+_47587129 | 0.06 |
ENST00000326656.8
ENST00000326674.9 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr17_+_21030260 | 0.05 |
ENST00000579303.1
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B |
| chr2_+_172543967 | 0.05 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr7_+_114562616 | 0.05 |
ENST00000448022.1
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr18_-_47017956 | 0.05 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr16_+_84875609 | 0.05 |
ENST00000563066.1
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr17_-_29151794 | 0.05 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr2_+_148778570 | 0.05 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr4_+_20255123 | 0.05 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr11_+_327171 | 0.05 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr22_+_40742512 | 0.05 |
ENST00000454266.2
ENST00000342312.6 |
ADSL
|
adenylosuccinate lyase |
| chr16_-_71264558 | 0.05 |
ENST00000448089.2
ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN
|
HYDIN, axonemal central pair apparatus protein |
| chr3_-_185542761 | 0.05 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_-_63815860 | 0.05 |
ENST00000272321.7
ENST00000431065.1 |
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr1_+_25599018 | 0.05 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr11_-_13484713 | 0.05 |
ENST00000526841.1
ENST00000529708.1 ENST00000278174.5 ENST00000528120.1 |
BTBD10
|
BTB (POZ) domain containing 10 |
| chr11_+_17298297 | 0.05 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr14_-_100841670 | 0.05 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr1_-_20834586 | 0.05 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr1_+_201798269 | 0.05 |
ENST00000361565.4
|
IPO9
|
importin 9 |
| chr6_+_127588020 | 0.05 |
ENST00000309649.3
ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146
|
ring finger protein 146 |
| chr6_+_10585979 | 0.05 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr10_-_12237820 | 0.05 |
ENST00000378937.3
ENST00000378927.3 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.2 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.2 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1900214 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0071810 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.0 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.0 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |