Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
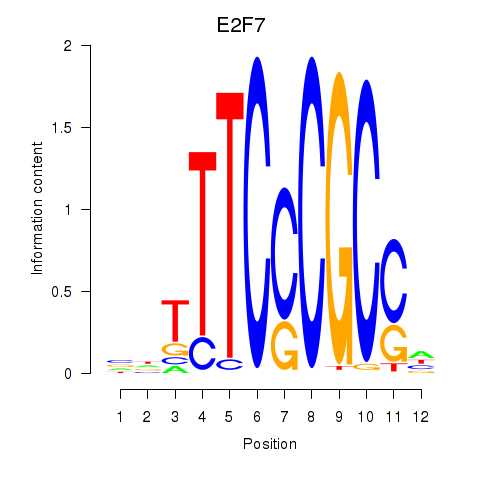
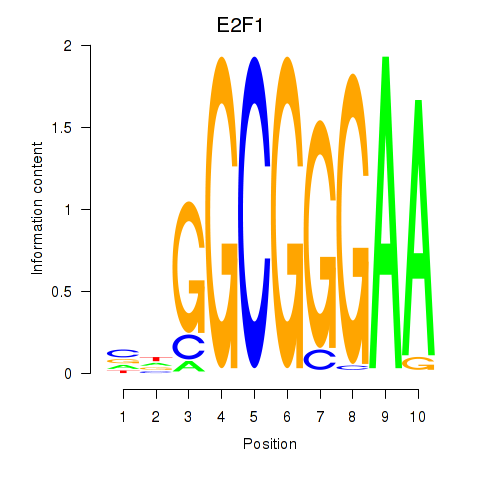
Results for E2F7_E2F1
Z-value: 2.03
Transcription factors associated with E2F7_E2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F7
|
ENSG00000165891.11 | E2F transcription factor 7 |
|
E2F1
|
ENSG00000101412.9 | E2F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F7 | hg19_v2_chr12_-_77459306_77459365 | 0.93 | 7.9e-03 | Click! |
| E2F1 | hg19_v2_chr20_-_32274179_32274213 | -0.27 | 6.0e-01 | Click! |
Activity profile of E2F7_E2F1 motif
Sorted Z-values of E2F7_E2F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48673465 | 5.37 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr1_+_60280458 | 1.61 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr19_+_48673949 | 1.50 |
ENST00000328759.7
|
C19orf68
|
chromosome 19 open reading frame 68 |
| chr8_+_103876528 | 1.23 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr10_+_102672712 | 1.16 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr13_+_42031679 | 1.11 |
ENST00000379359.3
|
RGCC
|
regulator of cell cycle |
| chr3_+_5163905 | 1.10 |
ENST00000256496.3
ENST00000419534.2 |
ARL8B
|
ADP-ribosylation factor-like 8B |
| chr15_+_44719996 | 1.10 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr5_+_68485433 | 1.08 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr12_+_72233487 | 1.07 |
ENST00000482439.2
ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15
|
TBC1 domain family, member 15 |
| chr17_-_58469591 | 1.07 |
ENST00000589335.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr11_-_72853267 | 1.03 |
ENST00000409418.4
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr14_+_38065052 | 1.00 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr15_+_44719970 | 0.97 |
ENST00000558966.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr1_+_51434357 | 0.97 |
ENST00000396148.1
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr6_-_17706618 | 0.93 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr18_-_658244 | 0.92 |
ENST00000585033.1
ENST00000323813.3 |
C18orf56
|
chromosome 18 open reading frame 56 |
| chr7_-_152373216 | 0.91 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr2_-_219433014 | 0.88 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr5_+_61602055 | 0.84 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr1_-_91487770 | 0.83 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr20_+_36661910 | 0.81 |
ENST00000373433.4
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr2_+_240323439 | 0.80 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr6_-_35656685 | 0.79 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr1_+_179263308 | 0.78 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr9_+_96928516 | 0.78 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr2_-_178129551 | 0.77 |
ENST00000430047.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr2_+_17935119 | 0.77 |
ENST00000317402.7
|
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr14_-_81902516 | 0.75 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr11_-_108369101 | 0.73 |
ENST00000323468.5
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2 |
| chr5_+_61601965 | 0.73 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr3_-_52719888 | 0.73 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr17_+_5390220 | 0.73 |
ENST00000381165.3
|
MIS12
|
MIS12 kinetochore complex component |
| chr22_+_46546406 | 0.72 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr5_+_61602236 | 0.71 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr16_+_19729586 | 0.71 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr2_-_17935059 | 0.71 |
ENST00000448223.2
ENST00000381272.4 ENST00000351948.4 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr5_+_68485363 | 0.70 |
ENST00000283006.2
ENST00000515001.1 |
CENPH
|
centromere protein H |
| chr7_-_100026280 | 0.69 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr15_+_44719790 | 0.69 |
ENST00000558791.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr18_+_29078131 | 0.68 |
ENST00000585206.1
|
DSG2
|
desmoglein 2 |
| chr11_+_9595180 | 0.68 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr1_+_6052700 | 0.67 |
ENST00000378092.1
ENST00000445501.1 |
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr3_+_37903432 | 0.67 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr10_-_70166999 | 0.67 |
ENST00000454950.2
ENST00000342616.4 ENST00000602465.1 ENST00000399200.2 |
RUFY2
|
RUN and FYVE domain containing 2 |
| chr16_+_81040794 | 0.67 |
ENST00000439957.3
ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN
|
centromere protein N |
| chr14_-_81902791 | 0.66 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr1_-_109825719 | 0.66 |
ENST00000369904.3
ENST00000369903.2 ENST00000429031.1 ENST00000418914.2 ENST00000409267.1 |
PSRC1
|
proline/serine-rich coiled-coil 1 |
| chr2_-_110371412 | 0.65 |
ENST00000415095.1
ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10
|
septin 10 |
| chr1_-_109825751 | 0.65 |
ENST00000369907.3
ENST00000438534.2 ENST00000369909.2 ENST00000409138.2 |
PSRC1
|
proline/serine-rich coiled-coil 1 |
| chr17_-_28618867 | 0.64 |
ENST00000394819.3
ENST00000577623.1 |
BLMH
|
bleomycin hydrolase |
| chr2_+_47630108 | 0.64 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr11_+_85956182 | 0.64 |
ENST00000327320.4
ENST00000351625.6 ENST00000534595.1 |
EED
|
embryonic ectoderm development |
| chr1_-_184943610 | 0.63 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr2_+_232575168 | 0.63 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr10_-_70166946 | 0.63 |
ENST00000388768.2
|
RUFY2
|
RUN and FYVE domain containing 2 |
| chr2_-_110371720 | 0.63 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr12_-_77459306 | 0.62 |
ENST00000547316.1
ENST00000416496.2 ENST00000550669.1 ENST00000322886.7 |
E2F7
|
E2F transcription factor 7 |
| chr14_-_45603657 | 0.62 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr5_+_126112794 | 0.61 |
ENST00000261366.5
ENST00000395354.1 |
LMNB1
|
lamin B1 |
| chr17_+_5389605 | 0.60 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr9_+_106856541 | 0.59 |
ENST00000286398.7
ENST00000440179.1 ENST00000374793.3 |
SMC2
|
structural maintenance of chromosomes 2 |
| chr6_-_35656712 | 0.59 |
ENST00000357266.4
ENST00000542713.1 |
FKBP5
|
FK506 binding protein 5 |
| chr7_-_158497431 | 0.59 |
ENST00000449727.2
ENST00000409339.3 ENST00000409423.1 ENST00000356309.3 |
NCAPG2
|
non-SMC condensin II complex, subunit G2 |
| chr6_-_18264706 | 0.59 |
ENST00000244776.7
ENST00000503715.1 |
DEK
|
DEK oncogene |
| chr3_+_128997798 | 0.58 |
ENST00000502878.2
ENST00000389735.3 ENST00000509551.1 ENST00000511665.1 |
HMCES
|
5-hydroxymethylcytosine (hmC) binding, ES cell-specific |
| chr11_-_82681626 | 0.58 |
ENST00000534396.1
|
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr18_+_43684298 | 0.57 |
ENST00000282058.6
|
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr4_+_184365841 | 0.57 |
ENST00000510928.1
|
CDKN2AIP
|
CDKN2A interacting protein |
| chr2_-_17935027 | 0.56 |
ENST00000446852.1
|
SMC6
|
structural maintenance of chromosomes 6 |
| chrX_-_20159934 | 0.56 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr13_+_34392200 | 0.56 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr1_+_91966384 | 0.56 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chr2_-_110371777 | 0.56 |
ENST00000397712.2
|
SEPT10
|
septin 10 |
| chr18_+_29077990 | 0.55 |
ENST00000261590.8
|
DSG2
|
desmoglein 2 |
| chr12_-_42538480 | 0.55 |
ENST00000280876.6
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr16_+_11439286 | 0.55 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr11_-_117186946 | 0.55 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr12_-_57472522 | 0.55 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr3_-_186524234 | 0.55 |
ENST00000418288.1
ENST00000296273.2 |
RFC4
|
replication factor C (activator 1) 4, 37kDa |
| chr3_-_186524144 | 0.54 |
ENST00000427785.1
|
RFC4
|
replication factor C (activator 1) 4, 37kDa |
| chr1_+_6845497 | 0.54 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr2_+_201936458 | 0.54 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr9_+_17135016 | 0.54 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr10_-_70092635 | 0.54 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr2_+_111878483 | 0.53 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr12_+_21654714 | 0.53 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr15_+_66585555 | 0.53 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr10_-_70092671 | 0.52 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr17_-_58469329 | 0.52 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr3_+_62304648 | 0.52 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr5_-_36242119 | 0.52 |
ENST00000511088.1
ENST00000282512.3 ENST00000506945.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr19_-_13030071 | 0.52 |
ENST00000293695.7
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chr1_-_243417762 | 0.51 |
ENST00000522191.1
|
CEP170
|
centrosomal protein 170kDa |
| chr7_-_30544405 | 0.50 |
ENST00000409390.1
ENST00000409144.1 ENST00000005374.6 ENST00000409436.1 ENST00000275428.4 |
GGCT
|
gamma-glutamylcyclotransferase |
| chr17_-_41277467 | 0.50 |
ENST00000494123.1
ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1
|
breast cancer 1, early onset |
| chr15_+_82555125 | 0.50 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr1_-_113162040 | 0.50 |
ENST00000358039.4
ENST00000369668.2 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr15_-_85259294 | 0.49 |
ENST00000558217.1
ENST00000558196.1 ENST00000558134.1 |
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr8_+_86019382 | 0.49 |
ENST00000360375.3
|
LRRCC1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr9_+_17134980 | 0.49 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr9_+_86595626 | 0.48 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr15_-_85259384 | 0.48 |
ENST00000455959.3
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr12_-_42538657 | 0.48 |
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr17_-_46724186 | 0.48 |
ENST00000433510.2
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr6_-_111804393 | 0.48 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr3_-_134204815 | 0.48 |
ENST00000514612.1
ENST00000510994.1 ENST00000354910.5 |
ANAPC13
|
anaphase promoting complex subunit 13 |
| chr14_+_36295504 | 0.48 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr15_-_59041768 | 0.48 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chrX_+_106871713 | 0.47 |
ENST00000372435.4
ENST00000372428.4 ENST00000372419.3 ENST00000543248.1 |
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr2_+_48757278 | 0.47 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr3_-_42003479 | 0.47 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr2_+_17935383 | 0.46 |
ENST00000524465.1
ENST00000381254.2 ENST00000532257.1 |
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr3_+_128997648 | 0.46 |
ENST00000509042.1
ENST00000383463.4 ENST00000417226.2 ENST00000510314.1 |
HMCES
|
5-hydroxymethylcytosine (hmC) binding, ES cell-specific |
| chr1_+_185014647 | 0.46 |
ENST00000367509.4
|
RNF2
|
ring finger protein 2 |
| chr17_+_34890807 | 0.46 |
ENST00000429467.2
ENST00000592983.1 |
PIGW
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr4_-_170679024 | 0.46 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr17_-_41277370 | 0.46 |
ENST00000476777.1
ENST00000491747.2 ENST00000478531.1 ENST00000477152.1 ENST00000357654.3 ENST00000493795.1 ENST00000493919.1 |
BRCA1
|
breast cancer 1, early onset |
| chr9_+_106856831 | 0.44 |
ENST00000303219.8
ENST00000374787.3 |
SMC2
|
structural maintenance of chromosomes 2 |
| chr5_+_93954039 | 0.44 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr2_+_7005959 | 0.44 |
ENST00000442639.1
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr15_-_49103235 | 0.44 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr13_+_49822041 | 0.43 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr5_+_14664762 | 0.43 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr18_-_54318353 | 0.43 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr1_-_36235529 | 0.43 |
ENST00000318121.3
ENST00000373220.3 ENST00000520551.1 |
CLSPN
|
claspin |
| chr2_-_102003987 | 0.43 |
ENST00000324768.5
|
CREG2
|
cellular repressor of E1A-stimulated genes 2 |
| chr1_+_91966656 | 0.42 |
ENST00000428239.1
ENST00000426137.1 |
CDC7
|
cell division cycle 7 |
| chr14_+_36295638 | 0.42 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr1_+_7844312 | 0.42 |
ENST00000377541.1
|
PER3
|
period circadian clock 3 |
| chr12_+_53443963 | 0.42 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr16_-_2205352 | 0.42 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr6_-_18155285 | 0.41 |
ENST00000309983.4
|
TPMT
|
thiopurine S-methyltransferase |
| chr13_+_34392185 | 0.41 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr6_-_18264406 | 0.41 |
ENST00000515742.1
|
DEK
|
DEK oncogene |
| chr7_-_127032363 | 0.41 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chrX_+_129473859 | 0.41 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_+_48010312 | 0.41 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6 |
| chr8_-_56685966 | 0.41 |
ENST00000334667.2
|
TMEM68
|
transmembrane protein 68 |
| chr12_+_53443680 | 0.41 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr4_-_39367949 | 0.41 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr6_-_146056341 | 0.40 |
ENST00000435470.1
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr2_+_48010221 | 0.40 |
ENST00000234420.5
|
MSH6
|
mutS homolog 6 |
| chr7_+_116593568 | 0.40 |
ENST00000446490.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr8_-_121457608 | 0.40 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr1_-_228401338 | 0.40 |
ENST00000295012.5
|
C1orf145
|
chromosome 1 open reading frame 145 |
| chr15_-_85259330 | 0.39 |
ENST00000560266.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr14_-_93673353 | 0.39 |
ENST00000556566.1
ENST00000306954.4 |
C14orf142
|
chromosome 14 open reading frame 142 |
| chr2_-_128145498 | 0.39 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr11_-_6341724 | 0.39 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr1_-_179851611 | 0.39 |
ENST00000610272.1
|
RP11-533E19.7
|
RP11-533E19.7 |
| chr1_+_149804218 | 0.39 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr2_-_235405679 | 0.39 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr11_+_85955787 | 0.39 |
ENST00000528180.1
|
EED
|
embryonic ectoderm development |
| chr8_+_48873479 | 0.38 |
ENST00000262105.2
|
MCM4
|
minichromosome maintenance complex component 4 |
| chr3_-_9811674 | 0.38 |
ENST00000411972.1
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr12_-_48499591 | 0.38 |
ENST00000551330.1
ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr11_+_17298297 | 0.38 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr19_-_409134 | 0.38 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C |
| chr15_-_49103184 | 0.38 |
ENST00000399334.3
ENST00000325747.5 |
CEP152
|
centrosomal protein 152kDa |
| chr4_+_17812525 | 0.38 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr2_-_54014127 | 0.38 |
ENST00000394717.2
|
GPR75-ASB3
|
GPR75-ASB3 readthrough |
| chr1_+_226250379 | 0.38 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr11_-_6341844 | 0.38 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chrX_+_146993534 | 0.38 |
ENST00000334557.6
ENST00000439526.2 ENST00000370475.4 |
FMR1
|
fragile X mental retardation 1 |
| chr9_+_80912059 | 0.37 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr2_-_136633940 | 0.37 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr14_+_100259666 | 0.37 |
ENST00000262233.6
ENST00000334192.4 |
EML1
|
echinoderm microtubule associated protein like 1 |
| chr2_+_219433588 | 0.37 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chrX_-_47509994 | 0.37 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chr10_-_126849626 | 0.36 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr15_-_85259360 | 0.36 |
ENST00000559729.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr7_-_21985489 | 0.36 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr14_-_61190754 | 0.36 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr16_+_2533020 | 0.36 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr4_+_83821835 | 0.36 |
ENST00000302236.5
|
THAP9
|
THAP domain containing 9 |
| chr16_-_18937726 | 0.36 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr5_+_43602750 | 0.36 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr16_-_72698834 | 0.36 |
ENST00000570152.1
ENST00000561611.2 ENST00000570035.1 |
AC004158.2
|
AC004158.2 |
| chr8_+_48873453 | 0.36 |
ENST00000523944.1
|
MCM4
|
minichromosome maintenance complex component 4 |
| chr5_-_146889619 | 0.36 |
ENST00000343218.5
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr2_-_239148599 | 0.35 |
ENST00000409182.1
ENST00000409002.3 ENST00000450098.1 ENST00000409356.1 ENST00000409160.3 ENST00000409574.1 ENST00000272937.5 |
HES6
|
hes family bHLH transcription factor 6 |
| chr1_+_95583479 | 0.35 |
ENST00000455656.1
ENST00000604534.1 |
TMEM56
RP11-57H12.6
|
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr1_+_225965518 | 0.35 |
ENST00000304786.7
ENST00000366839.4 ENST00000366838.1 |
SRP9
|
signal recognition particle 9kDa |
| chr14_-_36990061 | 0.35 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr11_+_17298255 | 0.35 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr6_+_116421976 | 0.35 |
ENST00000319550.4
ENST00000419791.1 |
NT5DC1
|
5'-nucleotidase domain containing 1 |
| chr2_-_176033066 | 0.35 |
ENST00000437522.1
|
ATF2
|
activating transcription factor 2 |
| chr15_+_66585879 | 0.35 |
ENST00000319212.4
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr4_-_174254823 | 0.34 |
ENST00000438704.2
|
HMGB2
|
high mobility group box 2 |
| chr2_-_46385 | 0.34 |
ENST00000327669.4
|
FAM110C
|
family with sequence similarity 110, member C |
| chr1_-_149832704 | 0.34 |
ENST00000392933.1
ENST00000369157.2 ENST00000392932.4 |
HIST2H4B
|
histone cluster 2, H4b |
| chr15_-_62352570 | 0.34 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr15_+_93426514 | 0.34 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr9_+_33524240 | 0.34 |
ENST00000290943.6
|
ANKRD18B
|
ankyrin repeat domain 18B |
| chr3_-_185826718 | 0.34 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr12_-_88535747 | 0.34 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chrX_-_63615297 | 0.34 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr17_+_76183398 | 0.34 |
ENST00000409257.5
|
AFMID
|
arylformamidase |
| chr2_-_24346218 | 0.34 |
ENST00000436622.1
ENST00000313213.4 |
PFN4
|
profilin family, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F7_E2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.4 | 1.2 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.4 | 1.2 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.4 | 1.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.3 | 0.9 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 | 1.2 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.3 | 0.9 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.3 | 0.9 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 1.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 0.7 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.2 | 1.9 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.8 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 0.6 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 | 1.2 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.2 | 0.5 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 4.7 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.1 | 0.9 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.6 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.5 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.8 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.1 | 0.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.8 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 1.4 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 1.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.6 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.9 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 1.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 0.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.3 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.5 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.3 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 0.3 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 0.3 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.5 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 1.8 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.4 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.6 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.3 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.6 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.2 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.1 | 0.6 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 1.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.1 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) |
| 0.1 | 0.3 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.3 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 0.2 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 0.3 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 1.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 2.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.4 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 0.4 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 1.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.2 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.7 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.3 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.4 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.6 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 0.4 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.1 | 0.7 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.4 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.4 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.2 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.2 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.2 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 1.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.4 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 0.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) seminal vesicle development(GO:0061107) |
| 0.1 | 1.4 | GO:0000732 | strand displacement(GO:0000732) |
| 0.1 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.3 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.7 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.3 | GO:0036260 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.2 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:0030099 | myeloid cell differentiation(GO:0030099) |
| 0.0 | 0.3 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.4 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.4 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.6 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.3 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.9 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0002351 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.3 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.3 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.0 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.9 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:1904438 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 1.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.5 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.3 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 1.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.1 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.4 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.1 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 1.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 1.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.9 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 2.2 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.0 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.2 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.0 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.3 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.2 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 1.2 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine(GO:0071233) cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.1 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:0032275 | luteinizing hormone secretion(GO:0032275) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:0070945 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) neutrophil mediated killing of gram-negative bacterium(GO:0070945) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) negative regulation of MHC class II biosynthetic process(GO:0045347) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.3 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.0 | 0.0 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.0 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.0 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.0 | GO:1903377 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0035194 | posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:2000146 | negative regulation of cell motility(GO:2000146) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.2 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.0 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.0 | GO:0044827 | modulation by host of viral genome replication(GO:0044827) |
| 0.0 | 0.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate biosynthetic process(GO:0030208) dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0003093 | regulation of glomerular filtration(GO:0003093) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 1.3 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.3 | 2.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 1.4 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 0.7 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.2 | 2.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 1.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 1.9 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.5 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.1 | 0.7 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.5 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.1 | 0.4 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 0.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.7 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.1 | 0.6 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.9 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 2.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.2 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.0 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.9 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.5 | GO:0035859 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 1.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.0 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 1.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.3 | 4.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 1.4 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.2 | 2.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.6 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.2 | 0.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.7 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.1 | 1.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.6 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.6 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.5 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.6 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.3 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 1.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.3 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 0.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 0.4 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.3 | GO:0031893 | vasopressin receptor binding(GO:0031893) V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 1.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.7 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.7 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.3 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.4 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.2 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.1 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.3 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 1.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 1.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.1 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.1 | 0.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.2 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.4 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.4 | GO:0048037 | cofactor binding(GO:0048037) |
| 0.0 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0016301 | kinase activity(GO:0016301) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.7 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 2.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.7 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 1.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 1.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.3 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.2 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 1.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 1.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 2.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 1.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.0 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.4 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.6 | GO:0016787 | hydrolase activity(GO:0016787) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 5.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 3.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.6 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 6.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.4 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.8 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |