Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
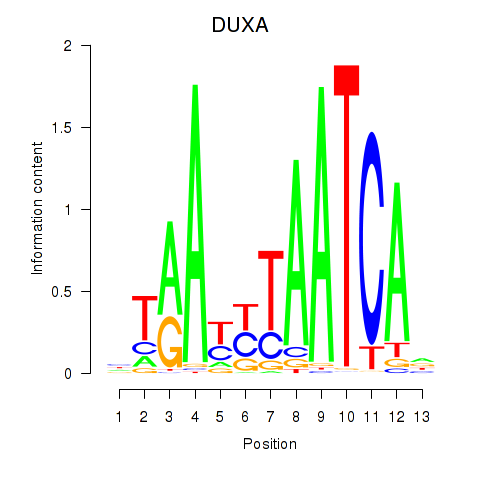
Results for DUXA
Z-value: 0.75
Transcription factors associated with DUXA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DUXA
|
ENSG00000258873.2 | double homeobox A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DUXA | hg19_v2_chr19_-_57678811_57678811 | 0.11 | 8.3e-01 | Click! |
Activity profile of DUXA motif
Sorted Z-values of DUXA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_228678550 | 1.14 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr1_-_153113927 | 0.68 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr15_-_80263506 | 0.66 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr22_-_30642728 | 0.53 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr6_-_133055896 | 0.48 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr6_-_133055815 | 0.39 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr22_+_31518938 | 0.39 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr13_-_30683005 | 0.38 |
ENST00000413591.1
ENST00000432770.1 |
LINC00365
|
long intergenic non-protein coding RNA 365 |
| chr11_+_93754513 | 0.36 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr12_-_49259643 | 0.33 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr4_-_112993808 | 0.32 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr1_-_27998689 | 0.32 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr12_+_27619743 | 0.28 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr2_-_175629164 | 0.27 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr19_-_57967854 | 0.26 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr18_+_61554932 | 0.24 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr12_+_120740119 | 0.24 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr1_+_44514040 | 0.24 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr4_-_87770416 | 0.23 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr1_-_12908578 | 0.23 |
ENST00000317869.6
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr7_+_134331550 | 0.23 |
ENST00000344924.3
ENST00000418040.1 ENST00000393132.2 |
BPGM
|
2,3-bisphosphoglycerate mutase |
| chr3_-_12587055 | 0.22 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chrX_+_154444643 | 0.22 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr10_-_112255945 | 0.22 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chr8_-_29605625 | 0.21 |
ENST00000506121.3
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr5_-_115872124 | 0.20 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr17_-_39553844 | 0.20 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chr10_+_118349920 | 0.20 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chrX_+_64708615 | 0.20 |
ENST00000338957.4
ENST00000423889.3 |
ZC3H12B
|
zinc finger CCCH-type containing 12B |
| chr2_+_201994042 | 0.20 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr9_-_19065082 | 0.19 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr12_-_51422017 | 0.19 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr18_-_59415987 | 0.18 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr10_+_69865866 | 0.18 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr8_-_91095099 | 0.17 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr1_+_104615595 | 0.17 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr18_+_44812072 | 0.17 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr1_+_218683438 | 0.17 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr9_-_75695323 | 0.17 |
ENST00000419959.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr20_+_58571419 | 0.16 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr6_+_89791507 | 0.16 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr1_+_41447609 | 0.16 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chrX_+_69501943 | 0.16 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr15_-_65407524 | 0.15 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr12_-_49581152 | 0.15 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr7_+_102196256 | 0.15 |
ENST00000341656.4
|
SPDYE2
|
speedy/RINGO cell cycle regulator family member E2 |
| chr12_-_111395610 | 0.15 |
ENST00000548329.1
ENST00000546852.1 |
RP1-46F2.3
|
RP1-46F2.3 |
| chrX_-_46759138 | 0.15 |
ENST00000377879.3
|
CXorf31
|
chromosome X open reading frame 31 |
| chr7_+_102295340 | 0.15 |
ENST00000455020.2
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr21_-_19858196 | 0.14 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr12_+_25348186 | 0.14 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr18_+_68002675 | 0.14 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr17_-_57158523 | 0.14 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr6_-_150346607 | 0.14 |
ENST00000367341.1
ENST00000286380.2 |
RAET1L
|
retinoic acid early transcript 1L |
| chr6_+_3259148 | 0.14 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr12_-_75784669 | 0.14 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr12_-_114211444 | 0.13 |
ENST00000510694.2
ENST00000550223.1 |
RP11-438N16.1
|
RP11-438N16.1 |
| chr12_+_25348139 | 0.13 |
ENST00000557540.2
ENST00000381356.4 |
LYRM5
|
LYR motif containing 5 |
| chr2_+_54342533 | 0.13 |
ENST00000406041.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr3_+_167582561 | 0.13 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr6_+_3259122 | 0.13 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr12_-_114211474 | 0.13 |
ENST00000550905.1
ENST00000547963.1 |
RP11-438N16.1
|
RP11-438N16.1 |
| chr16_-_18911366 | 0.12 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr14_+_20937538 | 0.12 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr7_-_34978980 | 0.12 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr11_-_114271139 | 0.12 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr19_-_47349395 | 0.12 |
ENST00000597020.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chrX_-_148571884 | 0.12 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr1_-_224624730 | 0.11 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr17_+_34842473 | 0.11 |
ENST00000490126.2
ENST00000225410.4 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chrX_+_133733457 | 0.11 |
ENST00000440614.1
|
RP11-308B5.2
|
RP11-308B5.2 |
| chr19_-_2090131 | 0.11 |
ENST00000591326.1
|
MOB3A
|
MOB kinase activator 3A |
| chr12_-_68835991 | 0.11 |
ENST00000546086.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr7_-_93520191 | 0.11 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr11_+_114270752 | 0.11 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr7_+_93690435 | 0.11 |
ENST00000438538.1
|
AC003092.1
|
AC003092.1 |
| chr3_+_118892411 | 0.11 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chrX_-_138072729 | 0.11 |
ENST00000455663.1
|
FGF13
|
fibroblast growth factor 13 |
| chr15_+_66679155 | 0.11 |
ENST00000307102.5
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr14_+_22600515 | 0.10 |
ENST00000390456.3
|
TRAV8-7
|
T cell receptor alpha variable 8-7 (non-functional) |
| chr11_+_5775923 | 0.10 |
ENST00000317254.3
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4 (gene/pseudogene) |
| chr16_-_21875424 | 0.10 |
ENST00000541674.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr16_-_57836321 | 0.10 |
ENST00000569112.1
ENST00000562311.1 ENST00000445690.2 ENST00000379655.4 |
KIFC3
|
kinesin family member C3 |
| chr1_+_153003671 | 0.10 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr1_+_209929494 | 0.10 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr7_-_93520259 | 0.10 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr3_+_118892362 | 0.10 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr5_-_115872142 | 0.10 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr21_-_23058648 | 0.10 |
ENST00000416182.1
|
AF241725.6
|
AF241725.6 |
| chr6_-_66417107 | 0.10 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr6_-_10838710 | 0.10 |
ENST00000313243.2
|
MAK
|
male germ cell-associated kinase |
| chr2_-_10587897 | 0.10 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr17_-_79849438 | 0.10 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr5_+_32585605 | 0.09 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr3_-_194188956 | 0.09 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr17_-_74733404 | 0.09 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr7_+_135611542 | 0.09 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr5_-_150537279 | 0.09 |
ENST00000517486.1
ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6
|
annexin A6 |
| chr3_-_120461353 | 0.09 |
ENST00000483733.1
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr11_-_18610246 | 0.09 |
ENST00000379387.4
ENST00000541984.1 |
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr1_+_152975488 | 0.09 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr11_+_62496114 | 0.09 |
ENST00000532583.1
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr4_+_147145709 | 0.09 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr15_-_34447023 | 0.09 |
ENST00000560310.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr5_+_172571445 | 0.09 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr8_-_125577940 | 0.09 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr1_-_198906528 | 0.08 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr6_+_12717892 | 0.08 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr16_+_57220193 | 0.08 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr12_-_110434096 | 0.08 |
ENST00000320063.9
ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr20_+_22034809 | 0.08 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr10_+_6821545 | 0.08 |
ENST00000436383.1
|
LINC00707
|
long intergenic non-protein coding RNA 707 |
| chr5_+_154237778 | 0.08 |
ENST00000523698.1
ENST00000517876.1 ENST00000520472.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr6_+_35996859 | 0.08 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr6_-_10838736 | 0.07 |
ENST00000536370.1
ENST00000474039.1 |
MAK
|
male germ cell-associated kinase |
| chr11_-_5526834 | 0.07 |
ENST00000380237.1
ENST00000396895.1 ENST00000380252.1 |
HBE1
HBG2
|
hemoglobin, epsilon 1 hemoglobin, gamma G |
| chr10_+_134150835 | 0.07 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr19_+_56187987 | 0.07 |
ENST00000411543.2
|
EPN1
|
epsin 1 |
| chr4_-_69536346 | 0.07 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr1_+_50459990 | 0.07 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr12_-_110434183 | 0.07 |
ENST00000360185.4
ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr4_-_106629796 | 0.07 |
ENST00000416543.1
ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12
|
integrator complex subunit 12 |
| chr2_-_224467002 | 0.07 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr15_+_78830023 | 0.07 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr16_+_57220049 | 0.07 |
ENST00000562439.1
|
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr6_+_86195088 | 0.07 |
ENST00000437581.1
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr3_-_33759699 | 0.07 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_+_45927994 | 0.07 |
ENST00000357632.2
ENST00000395963.2 |
CCR9
|
chemokine (C-C motif) receptor 9 |
| chr8_-_130587237 | 0.07 |
ENST00000520048.1
|
CCDC26
|
coiled-coil domain containing 26 |
| chr18_+_61445007 | 0.07 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr5_-_16742330 | 0.07 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chrX_+_100878079 | 0.07 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr8_-_86290333 | 0.07 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr11_-_6704513 | 0.07 |
ENST00000532203.1
ENST00000288937.6 |
MRPL17
|
mitochondrial ribosomal protein L17 |
| chr2_-_175629135 | 0.07 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr2_-_207078086 | 0.07 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr2_+_201994208 | 0.07 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_-_232651312 | 0.07 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr19_+_29456034 | 0.07 |
ENST00000589921.1
|
LINC00906
|
long intergenic non-protein coding RNA 906 |
| chr9_+_136325149 | 0.07 |
ENST00000542192.1
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr17_+_61271355 | 0.06 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr8_+_24151553 | 0.06 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr5_-_89705537 | 0.06 |
ENST00000522864.1
ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3
|
centrin, EF-hand protein, 3 |
| chr6_-_73935163 | 0.06 |
ENST00000370388.3
|
KHDC1L
|
KH homology domain containing 1-like |
| chrX_+_135614293 | 0.06 |
ENST00000370634.3
|
VGLL1
|
vestigial like 1 (Drosophila) |
| chr5_-_152069089 | 0.06 |
ENST00000506723.2
|
AC091969.1
|
AC091969.1 |
| chr6_+_31126291 | 0.06 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr18_+_19192228 | 0.06 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr4_+_79567057 | 0.06 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr5_+_154238149 | 0.06 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr12_-_90049828 | 0.06 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr6_-_71666732 | 0.06 |
ENST00000230053.6
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr5_+_67535647 | 0.06 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_-_76817036 | 0.06 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr21_+_47013566 | 0.06 |
ENST00000441095.1
ENST00000424569.1 |
AL133493.2
|
AL133493.2 |
| chr8_-_123440779 | 0.06 |
ENST00000533992.1
|
RP11-94A24.1
|
RP11-94A24.1 |
| chr6_+_53883708 | 0.06 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr5_-_75919253 | 0.06 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr5_+_125758865 | 0.06 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr12_-_110434021 | 0.06 |
ENST00000355312.3
ENST00000551209.1 ENST00000550186.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr7_+_18330035 | 0.06 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chr4_-_111563076 | 0.06 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr6_-_74104856 | 0.06 |
ENST00000441145.1
|
OOEP
|
oocyte expressed protein |
| chr17_-_5321549 | 0.06 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr1_-_154941350 | 0.06 |
ENST00000444179.1
ENST00000414115.1 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr11_-_11374904 | 0.06 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr2_+_198380763 | 0.06 |
ENST00000448447.2
ENST00000409360.1 |
MOB4
|
MOB family member 4, phocein |
| chr12_-_118628315 | 0.06 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr12_-_49245936 | 0.06 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr5_+_125758813 | 0.06 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr11_-_123525289 | 0.06 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr2_-_198062758 | 0.06 |
ENST00000328737.2
|
ANKRD44
|
ankyrin repeat domain 44 |
| chr4_-_89951028 | 0.05 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr1_-_21377383 | 0.05 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr5_-_75919217 | 0.05 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr17_+_34842512 | 0.05 |
ENST00000588253.1
ENST00000592616.1 ENST00000590858.1 ENST00000588357.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr15_-_43398274 | 0.05 |
ENST00000382177.2
ENST00000290650.4 |
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr1_-_26633480 | 0.05 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr13_+_76362974 | 0.05 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr2_+_86333340 | 0.05 |
ENST00000409783.2
ENST00000409277.3 |
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr13_-_76111945 | 0.05 |
ENST00000355801.4
ENST00000406936.3 |
COMMD6
|
COMM domain containing 6 |
| chr19_-_19302931 | 0.05 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr5_+_154238042 | 0.05 |
ENST00000519211.1
ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr20_-_44600810 | 0.05 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr1_+_74701062 | 0.05 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr12_+_31812121 | 0.05 |
ENST00000395763.3
|
METTL20
|
methyltransferase like 20 |
| chr17_+_7476136 | 0.05 |
ENST00000582169.1
ENST00000578754.1 ENST00000578495.1 ENST00000293831.8 ENST00000380512.5 ENST00000585024.1 ENST00000583802.1 ENST00000577269.1 ENST00000584784.1 ENST00000582746.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr16_+_67226019 | 0.05 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding |
| chr15_-_42749711 | 0.05 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr8_+_67104323 | 0.05 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr5_+_154238096 | 0.05 |
ENST00000517568.1
ENST00000524105.1 ENST00000285896.6 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr10_+_78078088 | 0.05 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr12_-_49245916 | 0.05 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr18_+_61445205 | 0.05 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_92632542 | 0.05 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr3_-_58419537 | 0.04 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr8_-_10512569 | 0.04 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr2_-_55237484 | 0.04 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr16_+_21623392 | 0.04 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr12_+_28299014 | 0.04 |
ENST00000538586.1
ENST00000536154.1 |
CCDC91
|
coiled-coil domain containing 91 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DUXA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.2 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.9 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.0 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0006710 | androgen catabolic process(GO:0006710) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 0.9 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |